Complete transcriptome evaluation of S. equorum pressure KM1031 beneath antibiotic stress
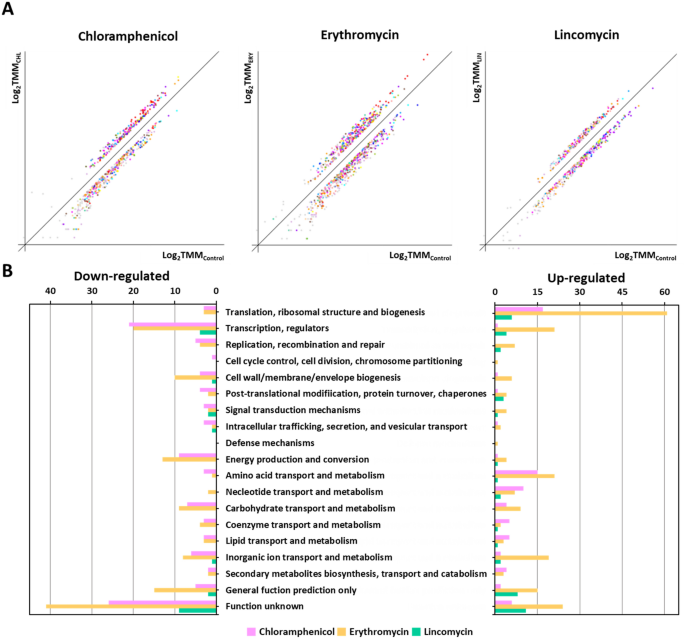
In a earlier examine, S. equorum pressure KM1031 confirmed resistance to chloramphenicol, erythromycin, lincomycin, and penicillin G primarily based on disk diffusion evaluation 10,13. Nonetheless, within the MIC outcomes, it was decided that pressure KM1031 was delicate to the penicillin G as a result of it didn’t develop at 1 mg/L of penicillin G. To grasp the bacterial response and diversifications throughout three antibiotic stress excluding penicillin G, RNA was remoted from S. equorum pressure KM1031 following antibiotic stress for RNA-Seq evaluation. RNA-Seq information have been acquired, mapped, and normalized as described within the “Strategies” part (Supplementary Tables S1 and S2). A complete of two,336 pressure KM1031 genes have been categorized utilizing COG evaluation. Antibiotic therapy affected the expression of a number of genes in pressure KM1031 (Supplementary Figs. S1 and S2). After mRNA abundance was in contrast between management and antibiotic-exposed cells, genes displaying a log2 (fold-change) larger than 2 or lower than − 2 have been thought of to be DEGs (Supplementary Tables S6 and S7; Supplementary Fig. S1). In pressure KM1031 cells uncovered to chloramphenicol, erythromycin, and lincomycin stress, 8.3% (183/2,336), 16.0% (354/2,336), and a couple of.9% (63/2,336) of genes exhibited vital variations of their expression, respectively (Fig. 1B).
Classification of differentially expressed genes (DEGs) primarily based on predicted features. (A) DEG evaluation from RNA-Seq information evaluating untreated Staphylococcus equorum pressure KM1031 with pressure KM1031 handled with antibiotics. The x-axis exhibits log-scaled Trimmed Imply of M-value (TMM) information for pressure KM1031, and the y-axis exhibits log-scaled TMM values for cells handled with chloramphenicol, erythromycin, and lincomycin, respectively. Complete gene expression within the two circumstances was filtered to determine considerably down- or upregulated genes with the factors P-value ≤ 0.05 and fold-change ≥ 2. (B) Genes upregulated or downregulated by twofold or extra following therapy of the bacterium with antibiotics have been grouped into purposeful classes primarily based on the Clusters of Orthologous Teams database.
Following chloramphenicol therapy, 75 genes have been considerably upregulated and 108 genes have been considerably downregulated (Supplementary Tables S6 and S7; and Fig. 1). Vital upregulation was noticed for genes related to translation, ribosomal construction, and biogenesis (22.7%; 17/75) in addition to genes related to amino acid transport and metabolism (20.0%; 15/75). In contrast, genes related to “operate unknown” (24.1%; 26/108) and transcription (19.4%; 21/108) have been downregulated.
Following erythromycin therapy, 214 genes have been considerably upregulated and 140 genes have been considerably downregulated. Vital upregulation was noticed for genes related to translation, ribosomal construction, and biogenesis (28.5%; 61/214) in addition to genes related to “operate unknown” (11.2%; 24/214). In contrast, genes related to “operate unknown” (29.3%; 41/140) and transcription (14.3%; 20/140) have been downregulated.
Following lincomycin therapy, 43 genes have been considerably upregulated and 20 genes have been considerably downregulated. Vital upregulation was noticed for genes related to “operate unknown” (25.6%; 11/43) in addition to genes related to translation, ribosomal construction, and biogenesis (14.0%; 6/43). In contrast, genes related to “operate unknown” (45.0%; 9/20) and transcription (20.0%; 9/20) have been downregulated.
Results of antibiotics on efflux proteins and transporters
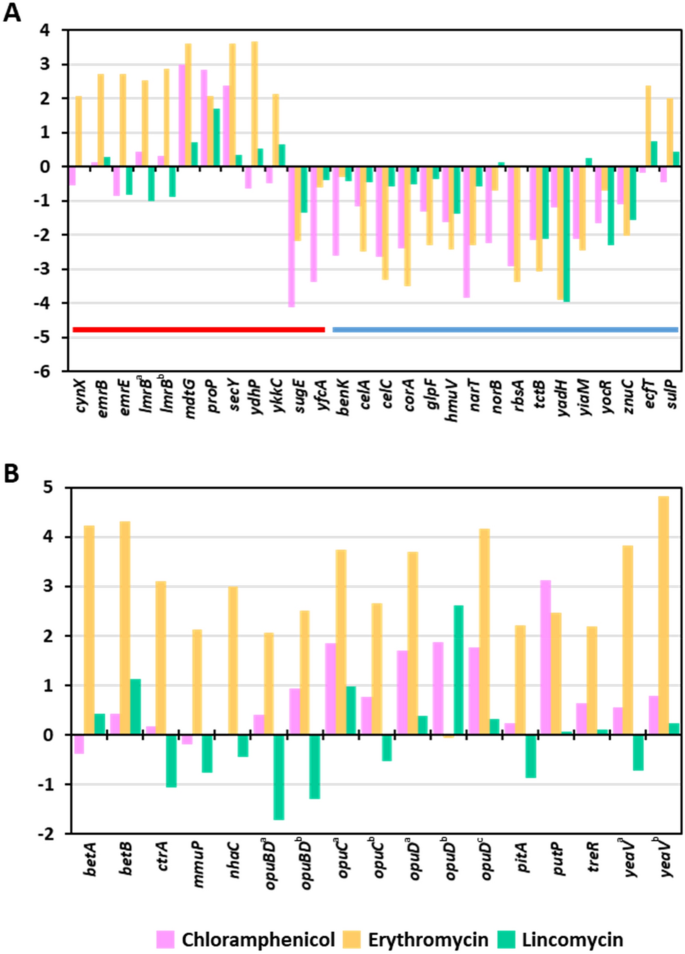
Transporter and efflux proteins are required for antibiotics to enter or be expelled from micro organism17,18. Thus, we hypothesized that antibiotics would alter the expression of efflux- and transporter-related genes in S. equorum. DEGs have been screened utilizing the key phrases “efflux” and “transporter.” Amongst DEGs following therapy with chloramphenicol, erythromycin and lincomycin, 7.1% (13/183), 6.8% (24/354), and 4.8% (3/63), respectively, have been associated to transporters and efflux (Fig. 2A; Supplementary Desk S8). Chloramphenicol and erythromycin therapy (particularly the previous) upregulated efflux-related genes and downregulated transporter-related genes. Related outcomes have been noticed for lincomycin, though expression adjustments have been much less dramatic in contrast with the opposite two antibiotics.
Results of antibiotics on expression of genes associated to salt tolerance
Accumulation or launch of appropriate solutes similar to glycine betaine, proline betaine, and carnitine confers salt tolerance by facilitating the response of cells to osmotic stress19. Curiously, osmoprotectant-related genes, similar to these concerned within the synthesis of trehalose, glycine betaine, choline, and proline, have been upregulated following chloramphenicol and erythromycin therapy (Fig. 2B; Supplementary Desk S8), whereas lincomycin solely barely affected the expression of some genes associated to salt tolerance. Zhu and Dai20 reported that overexpression of efflux pumps required for salt tolerance led to decreased antibiotic susceptibility. Our outcomes recommend that pressure KM1031 specific salt tolerance-related genes to counter the consequences of antibiotics, particularly chloramphenicol and erythromycin.
Responsive genes to 3 antibiotics primarily based on transcriptomic and comparative genomic analyses
We hypothesized that some genes in S. equorum pressure KM031 may be particularly and functionally (i.e., mechanistically) associated to chloramphenicol and erythromycin resistance. To determine such genes, we undertook comparative genomic evaluation of strains KM1031 (CRERLR), C2014 (CSESLS), and KS1039 (CSESLS).
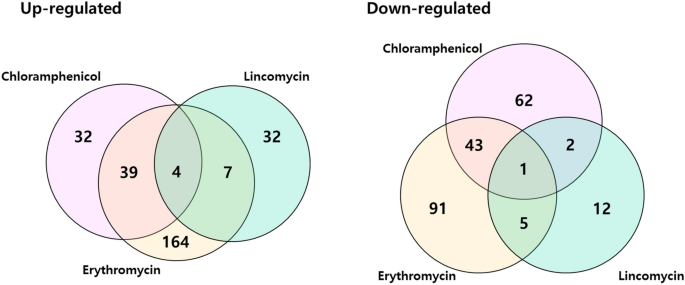
We plotted Venn diagrams of genes that have been considerably differentially expressed in S. equorum pressure KM031 in response to chloramphenicol, erythromycin and lincomycin (Fig. 3). 4 genes (AWC34_RS06585, AWC34_RS08650, AWC34_RS10220, and AWC34_RS12080) have been upregulated by all three antibiotics, whereas one (AWC34_RS11270) was downregulated by all three antibiotics (Supplementary Tables S6 and S7). These genes have been detected in a number of of the whole genome sequences of the antibiotic-sensitive S. equorum strains C2014 and KS1039 primarily based on comparative genomic evaluation.
Venn diagram of differentially expressed genes (DEGs) of S. equorum pressure KM1031 following therapy with chloramphenicol, erythromycin and lincomycin. Overlapping areas symbolize genes that have been differentially expressed in pressure KM1031 (in contrast with untreated cells) on therapy with two or three of the antibiotics. The numbers outdoors overlapping areas point out the numbers of considerably differentially expressed genes that have been affected by every antibiotic individually.
Curiously, an antibiotic ABC transporter ATP-binding protein-encoding gene (msr, AWC34_RS11115) was recognized amongst genes particularly upregulated in response to erythromycin. Msr is annotated, amongst different issues, as an erythromycin resistance ATP-binding protein. This gene was instructed to be chargeable for erythromycin resistance in a earlier genomic examine of S. equorum pressure KM103113. Reynolds et al. reported that Msr offers rise to erythromycin resistance through an lively transport course of21. Collectively, these findings strongly recommend that the antibiotic ABC transporter ATP-binding protein-coding gene (AWC34_RS11115) confers erythromycin resistance in pressure KM1031. Chloramphenicol- and lincomycin-specific response genes weren’t recognized amongst DEGs. Subsequently, we conclude that mostly up- and downregulated genes beneath antibiotic stress are related to normal environmental responses, and never responses to chloramphenicol, erythromycin, and/or lincomycin particularly.
We hypothesized that antibiotic publicity may improve the expression of genes which might be particularly associated to antibiotic resistance (i.e., that encode proteins which might be concerned within the molecular-level resistance of the micro organism to the drug). Thus, we took the set of genes that have been upregulated in S. equorum pressure KM031 (not DEGs) in response to any of the antibiotics and subtracted genes detected within the two CSESLS strains. This left 65 pressure KM1031-specific genes (Desk 1). The msr (AWC34_RS11115) gene was amongst them. In a earlier examine, we instructed that an antibiotic biosynthesis monooxygenase-encoding gene (abm AWC34_RS01805) and a lincosamide nucleotydyltransferase-encoding gene (lnuA, AWC34_RS13300) may confer resistance to chloramphenicol and lincomycin, respectively13. These genes have been additionally among the many KM1031-specific genes and abm and lnuA have been barely upregulated by publicity to chloramphenicol and lincomycin, respectively (Desk 1). The lincomycin-resistance phenotype of lnuA in pressure KM1031 has already been reported22. These outcomes suggest that abm could confer resistance to chloramphenicol, though abm was not considerably upregulated by chloramphenicol therapy.
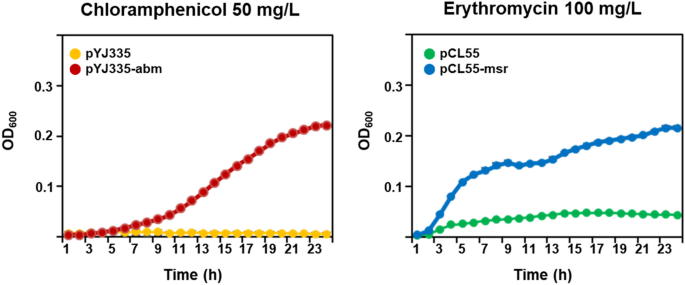
To research the impact of the abm and msr genes on chloramphenicol and erythromycin resistance, the genes AWC34_RS01805 and AWC34_RS11115 have been PCR amplified after which cloned into the pYJ335 and pCL55 vectors, respectively. The ensuing plasmids have been designated pYJ335-abm for the gene AWC34_RS01805 and pCL55-msr for the gene AWC34_RS11115. E. coli transformants harboring pYJ335-abm and pCL55-msr grew beneath chloramphenicol and erythromycin stress, respectively (Fig. 4). Collectively, these outcomes instructed that chloramphenicol and erythromycin therapy modified the expression of the abm and msr genes in S. equorum pressure KM031, and that the gene merchandise encoded by these genes contributed to the phenotypic resistance of E. coli cells to those antibiotics.
Results of antibiotics on two-component programs
Though we recognized particular genes that could be chargeable for the noticed antibiotic resistance of S. equorum pressure KM031, the transcriptional regulators of particular antibiotic resistance gene expression remained unclear. Two-component programs (TCSs) are the most typical programs for bacterial sign transduction in response to environmental alerts similar to antibiotics and salts. TCS signaling is concerned in bacterial resistance to antibiotics. A number of TCSs have been detected in S. equorum genomes13. Nonetheless, most TCS genes weren’t markedly up- or down-regulated in our experiments, besides the WalKR TCS (Desk 2). Though walKR genes weren’t considerably differentially expressed, expression of those genes elevated following therapy with chloramphenicol, erythromycin, and lincomycin. The WalKR TCS regulates genes chargeable for cell wall metabolism and homeostasis, in addition to genes concerned in stress responses, virulence, and biofilm formation23,24,25,26. Though the WalR consensus binding website (5′-TGTWAH N5 TGTWAH-3′)27 was not recognized upstream of abm, msr or lnuA, we hypothesize that the WalKR TCS may be associated to expression of those three antibiotic-specific responsive genes.
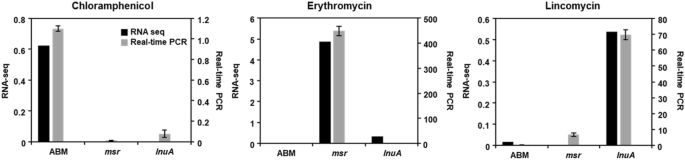
Validation of RNA-Seq information by qRT-PCR
qRT-PCR was used to validate the S. equorum pressure KM031 transcriptional profiles obtained by RNA-Seq evaluation. As proven in Fig. 5, the expression patterns for every gene (abm, msr, and lnuA) have been comparable by qRT-PCR and RNA-Seq. Expression of the abm, msr and lnuA genes elevated following publicity to chloramphenicol, erythromycin, and lincomycin, respectively. As well as, expression of walKR genes was elevated following publicity the three antibiotics, though not considerably (Supplementary Fig. S3). Thus, our RNA-Seq information have been confirmed by qRT-PCR.
Validation of RNA sequencing information by quantitative real-time PCR (qRT-PCR). Genes associated to resistance to chloramphenicol, erythromycin, and lincomycin have been chosen for validation beneath completely different antibiotic pressures. Knowledge are expressed as log2 fold-changes in gene expression between management and antibiotic-treated samples. In qRT-PCR, 16S rRNA gene expression was used for normalization of goal gene expression.