The affect of the TDA-producer P. inhibens DSM 17395 (P. inhibens (WT)), and its TDA-deficient mutant, on the microbial group related to the marine microalgae T. suecica was decided by sequencing of 16S rRNA gene V3-V4 area amplicons and evaluating the taxonomic range, composition, and construction over 70 days (Fig. 1A). In complete, 46 bacterial ASVs had been noticed within the microalgal microbiome throughout the whole time course. Out of those, 29 ASVs had been distributed on 20 genera, excluding Phaeobacter, represented 95.1% of the overall microbiome over the whole research (Fig. 1B). Of those, the highest 5 genera had been Jejuia, SM1A02 (household: Phycisphaeraceae), Algoriphagus, Marivita and Hoeflea with imply relative abundances of 33.1 ± 13.2%, 18.8 ± 9.0%, 9.9 ± 4.9%, 8.2 ± 8.8% and three.7 ± 4.3%, respectively (Fig. S1).
The preliminary abundance of T. suecica was decided each seventh day and used as an indicator for system well being and stability [4]. After inoculation with log 4.5 ± 0.1 cells mL−1, T. suecica abundance elevated to log 6.0 ± 0.1 cells mL−1 throughout the two weeks of development (Fig. 1C). Following the bi-weekly switch, T. suecica regrew to this degree, with no important distinction between the three methods (p = 0.11, LMM), indicating that the microalgal host was unaffected by the addition of P. inhibens (WT) or the TDA-deficient mutant, thus guaranteeing a steady mannequin system.
The power of Phaeobacter to provide TDA is a minor driver of range
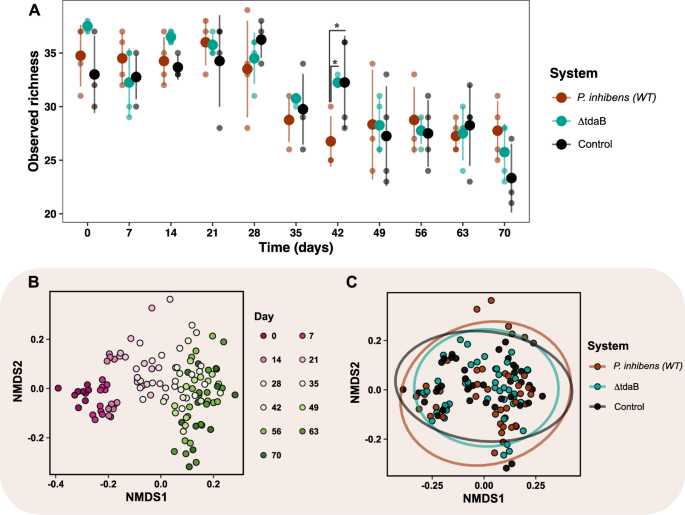
If P. inhibens makes use of TDA as an antagonistic compound to kill rivals, a lower in bacterial richness could possibly be anticipated within the methods uncovered to TDA-producing P. inhibens (WT). Due to this fact, the impact of P. inhibens strains on ASV richness of the bacterial communities was estimated as variety of noticed ASVs over time (Fig. 2A). No important distinction in richness was discovered between the three methods over time (p = 0.86, LMM), besides from day 42, the place the addition of P. inhibens (WT) triggered a major decrease richness than discovered within the different two methods (p = 0.017, EMMs). The variations on day 42 was brought on by ASVs belonging to the genera Halomonas, Haliea, Muricauda, Roseobacter and Methylophage not being detected within the P. inhibens (WT) system (Fig. S3). Nevertheless, the variety of noticed ASVs was considerably completely different over time in all methods (p < 2e–16, LMM), defined by the noticed richness decreased with 9.5 ± 2.4 ASVs from day zero to 70. The parallel discount in richness signifies that different elements, such because the biweekly dilution of the methods, moderately than the addition of the TDA-producing P. inhibens (WT) or a TDA-deficient mutant, drove this richness discount over time.
A Noticed richness (alpha range depicted as variety of ASVs) of the three methods in response to the addition of Phaeobacter inhibens DSM17395 (WT), TDA-deficient P. inhibens DSM17395 (ΔtdaB) and no addition (Management) over 70 days. Asterisks signify important variations (p < 0.05, LMM & EMM). Stuffed circles signify averages and clear factors organic replicates (lineages). Error bars signify commonplace deviation (N = 4). B Non-metric multidimensional scaling (NMDS) plot on Bray–Curtis distances of ASV composition. The primary two of three dimensions are proven. Factors signify samples which can be coloured in accordance with time, and C system. The ellipses signify 95% confidence interval. Stress = 0.09986.
Variables influencing the microbial group composition
Though P. inhibens (WT) didn’t have robust affect on the presence/absence of ASVs within the T. suecica microbiome, TDA-producers could possibly be chargeable for modulating and altering the expansion of group members. We used NMDS ordination to visualise Bray–Curtis dissimilarities of communities, and permutational multivariate evaluation of variance (PERMANOVA) to check for important variations. We excluded ASVs labeled as Phaeobacter from this evaluation to concentrate on the impact of Phaeobacter. The PERMANOVA revealed that 37.95% of the variation noticed within the bacterial composition had been defined by time and kind of system (Fig. 2B, C). The bacterial communities segregated in accordance with time (PERMANOVA: R2 = 0.344, p = 0.001; beta-dispersion: p > 0.05), which defined 34.4% of the variation noticed. This sample was additionally noticed within the ordination visualization, the place a transparent division between days is noticed from day zero to twenty-eight, adopted by a much less clear temporal sample from day 35 to 70 (Fig. 2B), suggesting diversification in all methods throughout time. The bacterial communities additionally segregated based mostly on the kind of system (PERMANOVA: R2 = 0.036, p = 0.001; beta-dispersion: p > 0.05), though this solely defined 3.6% of the variation, which was not visually evident within the ordination visualization (Fig. 2C), nor when choosing different dimensions (Fig. S5B). Nevertheless, multilevel pairwise PERMANOVA between the P. inhibens (WT) and the TDA-deficient mutant methods revealed that the group composition was influenced by the flexibility to provide TDA (PERMANOVA: R2 = 0.024, p = 0.003). This factors to the truth that the presence of P. inhibens (WT) was solely a minor driver of shaping the group composition.
We speculated that the small impact of the P. inhibens (WT) could possibly be defined by every organic replicate (lineage) having completely different evolutionary trajectories, doubtlessly owing to stochasticity derived from small variations in beginning situations. Certainly, the PERMANOVA revealed that lineage may clarify 15.6% of the variance noticed (PERMANOVA: R2 = 0.156, p = 0.001; beta-dispersion: p > 0.05), and that the impact of lineage relied on time (PERMANOVA: R2 = 0.079, p = 0.001; beta-dispersion: p > 0.05). We, due to this fact, tracked the trajectories of the bacterial group composition of every lineage (Fig. S6), which revealed that particularly lineage three within the P. inhibens (WT) system diversified from the opposite P. inhibens (WT) lineages from day 21 to 70. This could possibly be defined by two ASVs, Marinobacter ASV25 (r = 0.59, p = 0.001) and Sulfitobacter ASV14 (r = 0.6, p = 0.001), having a relative larger abundance, and Marivita ASV5 (r = 0.58, p = 0.001) having a relative decrease abundance at as of late in comparison with the remaining P. inhibens (WT) system lineages (Desk S3). This indicated that lineages throughout the three methods exhibited distinct temporal patterns in group composition.
Relative abundance trajectories of single ASVs reveal {that a} subset of ASVs is influenced by the presence of Phaeobacter
Since we noticed temporal variability between lineages throughout the similar system (Fig. S6), elucidating variations in ASVs trajectories between methods may be troublesome. Due to this fact, to make clear the underlying dynamics within the microbial communities, we used linear combined impact modeling (random impact being the lineage) to determine important relative abundance trajectories of ASVs between the three methods, solely together with ASVs that had been current in multiple pattern.
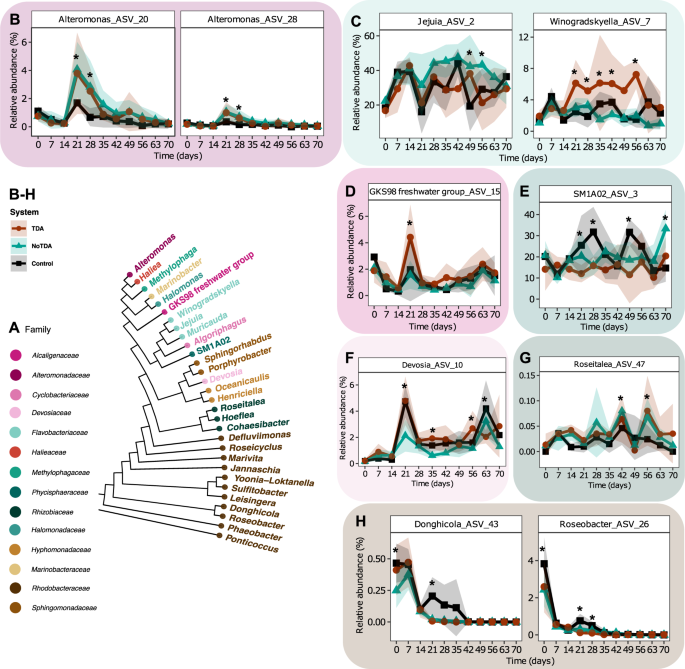
Out of those 40 ASVs (Fig. S7), ten ASVs displayed important completely different trajectories between the three methods no less than one timepoint (p < 0.05, EMMs, Fig. 3B–H, Desk S4). After the primary batch switch at day 21 to twenty-eight, the 2 Alteromonas ASVs 20 and 28 gained a major larger relative abundance within the methods with the P. inhibens (WT) (day 21: p = 0.005 and p = 0.001; day 28: p = 0.0004 and p = 0.001, EMMs) and the TDA-deficient mutant (day 21: p = 0.014 and p = 0.005; day 28: p = 0.002 and p = 0.009, EMMs) in comparison with the management system (Fig. 3B), indicating that the upper relative abundances of Alteromonas ASVs had been primarily an impact of the addition of P. inhibens, moderately than the flexibility of the bacterium to provide TDA. In distinction, an ASV labeled as Donghicola (ASV43) disappeared quicker from the 2 P. inhibens methods after day 21 (WT: p < 0.0001 and TDA-deficient mutant: p < 0.0001, in comparison with the management system, EMMs), whereas the ASV stays within the management system till day 35 earlier than it goes extinct (Fig. 3H). Moreover the earlier than talked about ASVs, 5 ASVs displayed important variations at day 21, together with Winogradskyella ASV7 (Fig. 3C), GKS98 freshwater group ASV15 (Fig. 3D), SM1A02 ASV3 (Fig. 3E), Devosia ASV10 (Fig. 3F), Roseitalea ASV47 (Fig. 3G) and Roseobacter ASV26 (Fig. 3H). Nevertheless, the variations at day 21 just isn’t related between ASVs (Fig. 3). These modified dynamics following day 21 point out that the batch switch resets the communities differentially relying on whether or not P. inhibens has been added beforehand to the methods. Curiously, the Winogradskyella ASV7 displayed larger relative abundance in communities with P. inhibens (WT) in comparison with the 2 different methods after the batch switch at day 21 till day 56 (Fig. 3C), the place some days the relative abundance was important completely different (p < 0.05, EMMs). This means that the Winogradskyella pressure is positively influenced by the presence of a TDA-producing Phaeobacter. Altogether, the addition of Phaeobacter, able to producing TDA or not, can affect the relative abundance trajectories of strains current within the microalgae group over time.
A Phylogenetic tree with recognized genus names of ASVs analyzed. Genera names are coloured by household. B–H Relative abundance trajectories of serious completely different ASVs, recognized utilizing linear combined impact fashions (LMM). Asterisks signify timepoints with important variations between two or extra methods (p < 0.05, LMM and EMM). Particular important variations are listed in Desk S4. Strong traces signify imply relative abundance and ribbons commonplace deviation of ASVs labeled at genus degree as B Alteromonas, C Jejunia, Winogradskyella, D GKS98 freshwater group, E SM1A02, F Devosia, G Roseitelea, and H Donghicola and Roseobacter (N = 4).
Absolutely the abundance profiles of Phaeobacter are completely different throughout methods
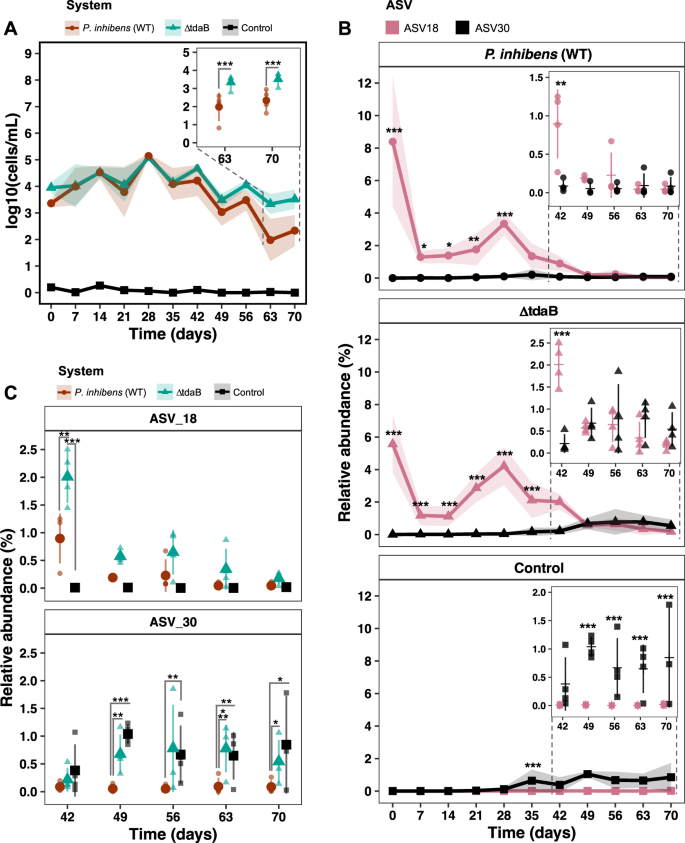
We speculated whether or not the earlier observations had been brought on by the elevated inoculum of Phaeobacter or by continued interplay over-time with Phaeobacter strains. We due to this fact sought to quantify the abundance of Phaeobacter by qPCR utilizing genus particular primers [18]. There was a major impact of time on absolutely the abundance of Phaeobacter (p < 2.2e–16, LMM), for the reason that degree of Phaeobacter elevated from log 3.4 ± 0.2 and three.95 ± 0.2 CFU mL−1 within the P. inhibens (WT) and TDA-deficient mutant system, respectively, at day zero to approx. log 5 CFU mL−1 at day 28, adopted by a lower to log 2–3 CFU mL−1 (Fig. 4A). Thus, the extent of Phaeobacter within the P. inhibens (WT) and TDA-deficient mutant system adopted related dynamics till day 56–70, the place the extent of Phaeobacter within the TDA-deficient mutant system was considerably larger than within the P. inhibens (WT) system (p < 0.05, EMMs). The qPCR strategy additionally detected a low degree of Phaeobacter (log 0.2 ± 0.26 CFU mL−1) being current within the management system at day zero, nonetheless, the abundance of this Phaeobacter inhabitants remained between log 0.2 ± 0.26 and 0 CFU mL−1, whereas ranges of Phaeobacter within the P. inhibens (WT) and TDA-deficient mutant methods remained larger (p < 0.0001, EMMs), indicating that each Phaeobacter strains established themselves within the system.
A Absolutely the abundances of micro organism belonging to the Phaeobacter genus measured by qPCR utilizing genus-specific primers. Symbols signify means and coloured ribbons the usual deviation. Asteriks denotes significance degree (p < 0.05, LMM and EMM). (N = 4). B Relative abundance of ASVs labeled as Phaeobacter at genus degree within the three methods. Factors signify means and coloured ribbons the usual deviation. Within the time subset plots, horizontal traces signify means, crammed symbols replicates and error bars the usual deviation. Asteriks denotes significance degree (p < 0.05, LMM and EMM). (N = 4). C Comparisons of the 2 Phaeobacter ASVs: ASV18 and ASV30 between the three methods. Stuffed symbols signify means, clear symbols replicates and error bars the usual deviation. Asteriks denotes significance degree (p < 0.05, LMM and EMM). (N = 4).
Expression of tdaC within the algal experimental setup
Variations in group composition had been seen between the methods uncovered to P. inhibens (WT) and the TDA-deficient mutant, nonetheless, it isn’t recognized if TDA is definitely produced within the system. Now we have beforehand not been in a position to detect TDA chemically in algal methods [17] and makes an attempt on this research had been additionally not profitable (knowledge not proven). We due to this fact determined to make use of gene expression as a proxy for the precise manufacturing and developed a RT qPCR protocol for detecting the expression of the tdaC gene. tdaC transcripts had been detected in three out of 4 replicates (684 ± 854 transcripts mL−1) within the algal cultures, to which the P. inhibens (WT) was added. tdaC transcripts had been additionally detected within the TDA-deficient mutant algal cultures in two out of 4 replicates (1137 and 2336 transcripts mL−1). The tdaC gene was detected in all P. inhibens (WT) and TDA-deficient mutant samples at day one, 4, and 7, and ranged from log 4.9–5.5 genes mL−1, day one being barely larger than day 4 and 7. The TDA-deficient mutant has an insert within the tdaB gene and doesn’t, in pure cultures, produce TDA. To make sure that the detection of tdaC transcripts within the algal cultures, to which we added the TDA-deficient mutant was not as a consequence of contamination, we PCR-amplified the tdaB gene from the samples and this was solely detected in algal cultures, to which the P. inhibens (WT) was added.
Indigenous Phaeobacter ASV beneficial properties footing when TDA-producing Phaeobacter usually are not current
Discovering micro organism belonging to the Phaeobacter genus within the management system led us to question whether or not these indigenous Phaeobacter had been additionally current within the manipulated methods, and in that case, at what ranges. Relating distinctive ASVs to particular bacterial strains, and even species, may be troublesome, if not deceptive [57]. Nevertheless, most species throughout the Phaeobacter genus may be distinguished by the 16S rRNA gene, as most species have distinct 16S alleles, together with P. inhibens [58]. Out of the 46 ASVs current within the communities, two ASVs had been labeled as Phaeobacter at genus degree (Fig. S8). The 2 Phaeobacter ASVs had been recognized as ASV18 matching P. inhibens, representing each the wildtype and the TDA-deficient mutant, and ASV30 matching one other Phaeobacter, which has two nucleotide substitutions relative to Phaeobacter ASV18 (Fig. S8).
The relative abundance of ASV18 was considerably completely different between the three methods, between days and the distinction between methods relied on the day (p < 2.2e–16, LMM, Fig. S7). The relative abundance profile of ASV18 from day zero to 35 had been larger within the manipulated methods in comparison with the management (p < 0.05, EMMs, Fig. 4B). At day 42, the relative abundance decreased within the P. inhibens (WT) system (Fig. 4C), leading to no important distinction between the P. inhibens (WT) and management system (p = 0.06, EMMs), however there was a considerably larger relative abundance of ASV18 within the TDA-deficient mutant system in comparison with each the P. inhibens (WT) and management (p = 0.01 and p = 2.1e–6, respectively, EMMs). Within the remaining timepoints (day 49 to 70), no important variations had been discovered between both of the methods (p > 0.05, EMMs, Fig. 4C).
There was additionally a major impact of time (p < 2.2e–16, LMM) on ASV30, and a major distinction between the methods (p = 0.008, LMM), which clearly relied on time (p = 2.1e–06, LMM, Fig. S7). The relative abundance dynamics of ASV30 had been completely different from that of ASV18 in all methods (Figs. S7 and 4B). From day zero to 42 the relative abundance of ASV30 had been under 1.6% within the management system, 0.6% within the TDA-deficient mutant system, and under 0.8% within the P. inhibens (WT), and due to this fact no important variations had been discovered between methods (p > 0.05, EMMs). From day 49 to 70, the relative abundances of ASV30 had been considerably decrease within the P. inhibens (WT) system in comparison with management system (p < 0.05, EMMs, Fig. 4C) and the TDA-deficient mutant system (p < 0.05, EMMs, Fig. 4C), besides from day 56, the place the extent of ASV18 was related between the TDA-deficient mutant and P. inhibens (WT) system (p = 0.06, EMMs, Fig. 4C). In distinction, no important variations had been discovered between the TDA-deficient mutant and management system in the identical interval (p > 0.05, EMMs, Fig. 4C), indicating that the relative abundance of ASV30 was the identical within the two methods.
The numerous variations noticed in absolute abundance of the Phaeobacter genus between the P. inhibens (WT) and TDA-deficient mutant system within the late part (Fig. 4A) could possibly be defined by a better relative abundance of ASV30 within the TDA-deficient mutant system (Fig. 4C). In reality, ASV18 co-existed with ASV30 for a interval within the TDA-deficient mutant system, a sample not noticed within the P. inhibens (WT) system (Fig. 4B, C). As compared, ASV30 was barely noticed within the P. inhibens (WT) system at any time level, therefore probably the most dominant Phaeobacter inhabitants may be assigned to ASV18 (Fig. 4B, C). Altogether these observations indicated that the addition of Phaeobacter (each P. inhibens (WT) and TDA-deficient mutant) suppressed the naturally occurring Phaeobacter.
Because the TDA-producing P. inhibens depleted quicker over time in comparison with the TDA-deficient mutant, one may speculate whether or not this pressure skilled a stronger choice strain. To find out if the selective strain was based mostly on mutational adjustments, we remoted 48 isolates of TDA-producing P. inhibens (WT) at day 35, 63, and 70 for complete genome sequencing. At as of late, we estimated that P. inhibens reached roughly 38.5, 69.3, and 75 generations, respectively. Nevertheless, we didn’t observe any constant mutation or sample that indicated related genetic adaptions to the microalgal group or host (Desk S5).