The current research printed in Science and Cell Analysis investigated the buildings and biochemical mechanism of the SARS-CoV-2 Omicron BA.1 and BA.2 spike trimer with ACE2 in addition to an efficient antibody JMB2002 in opposition to BA.1 and BA.2 spike, which offer new insights for the worldwide improvement of broad-spectrum anti-SARS-CoV-2 antibodies and vaccines.1,2
The emergent extreme acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant Omicron (BA.x) and its sublineages with enhanced immune escape capability have posed hostile influence and demanding challenges to the efficacy of neutralizing antibody therapeutics and SARS-CoV-2 vaccines. For the reason that Omicron variant emerged in Africa in 2021, it quickly turns into the dominant variant, changing the previous Delta and different variants all over the world. Omicron BA.1 subvariant emergent in November 2021 was quickly changed by one other Omicron subvariant BA.2 in early 2022. Nonetheless, two subsequent emergent subvariants, BA.4/BA.5, discovered first in Africa with further L452R and F486V mutations, competed with the previous subvariant BA.2 with enhanced infectivity and antibody evasion capability3,4 (Fig. 1a, b).
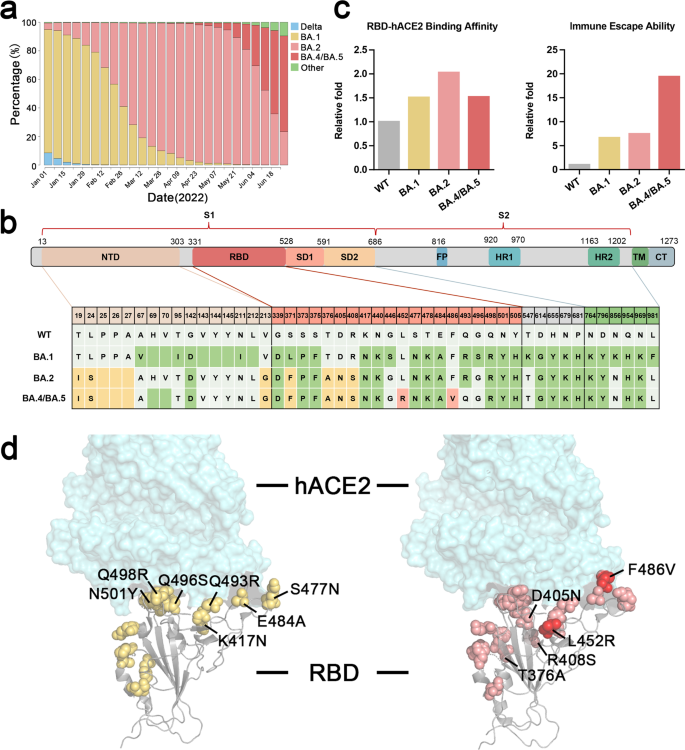
Comparability of prevalence proportion, construction and mutations, binding affinity, and immune escape capability of Omicron subvariants BA.1, BA.2, and BA.4/5. a Prevalence proportion of Omicron subvariants since January 1, 2022 (Variant distribution knowledge can be found in https://covidcg.org/). b The mutations within the spike protein of Omicron subvariants BA.1, BA.2, and BA.4/BA.5, that are marked in several colours (inexperienced for brand spanking new mutations in BA.1, yellow for brand spanking new mutations in BA.2, pink for brand spanking new mutations in BA.4/BA.5). c Comparability of RBD-hACE2 relative binding affinity and immune escape capability amongst Wildtype (WT), BA.1, BA.2, and BA.4/BA.5. Information of binding affinity and immune escape capability check with the reported wet-bench knowledge.3,4 d Construction and mutation annotation of RBD binding with hACE2. Yellow for mutations in Omicron BA.1, pink for mutations in BA.2, and pink and crimson for mutations in BA.4/BA.5
Omicron variants carry great amount of mutations on the floor of the spike protein (>30 amino acid mutations), which have raised involved of altering binding epitopes and inflicting escapes from nearly all of current SARS-CoV-2 therapeutical antibodies. Omicron BA.1 RBD varieties further interactions with ACE2 in comparison with wildtype strains (WT) because of the RBD mutations of S477N, Q493R, Q496S, Q498R, and N501Y in addition to the mutations K417N and E484A compensating the lack of polar interactions (Fig. 1b).
Based on the research, the RBD-hACE2-binding affinity of Omicron BA.1 and BA.4/BA.5 is about 1.5-fold greater than WT, whereas BA.2 is about twofold greater than WT.1,2,3,4 (Fig.1c). With regard to the spike trimer, Omicronbound ACE2 with an elevated affinity of about 6.2-folds (BA.1) and 11.1-folds (BA.2) in contrast with the WT spike trimer.1,2 BA.2 spike trimer certain ACE2 with an elevated affinity of about 1.8-folds in contrast with BA.1. The weird RBD-RBD interplay mechanism revealed that RBD within the Omicron spike trimer was stabilized in open-up conformation, which contributed to the upper affinity of Omicron variants. This may occasionally play an necessary function in its greater infectivity in contrast with former variants.1 Though the RBD of BA.1 spike trimer certain just one hACE2, Omicron BA.2 spike trimer may bind at the least two or three hACE2, which was noticed by cryo-EM structural evaluation and indicated a stronger hACE2-binding capability of the BA.2 spike trimer.2
Within the examine printed in Cell Analysis, the excessive binding effectivity between Omicron spike trimer and mouse ACE2 was discovered, in comparison with WT spike trimer from pressure Wuhan-Hu-1.2 Nonetheless, each Omicron and WT spike trimers hold excessive binding effectivity with cat ACE2. These knowledge counsel potential zoonotic occasions from Omicron variants and transmissible dangers between contaminated human and animals.
Most Omicron mutations had been discovered on the floor of the spike protein, a lot of that are the identified epitopes focused by the therapeutic antibodies. There are 15–17 mutations emergent within the Omicron RBD, which comprise the receptor-binding websites and the epitopes for the foremost antibodies induced by infections or vaccinations. Earlier research have raised the priority of enhanced immune escape capability from vaccinated sera and antibodies in opposition to Omicron variants3,4 (Fig.1c, d).
The RBD of newly emerged BA.4 and BA.5 subvariants encompasses the altered epitopes inflicting therapeutic antibody escape, e.g., Bril-196 of sophistication 1, REGN10933 and LYCoV555 of Class 2, and REGN10987 and Bril-198 of Class 3.1,3 A broad-spectrum therapeutic antibody, JMB2002 which may neutralize numerous variants together with Alpha, Beta, and Gamma, has accomplished the scientific trial of Part I. It may additionally neutralize Omicron BA.1 and BA.2. JMB2002 Fab certain the Omicron spike trimer with elevated affinity of BA.1 (OkD = 3.2 ± 3.0 nM) and BA.2 (roughly 2.6 nM) in contrast with the WT spike trimer (OkD = 12.2 ± 11.6 nM). In the meantime, JMB2002 IgG certain the Omicron BA.1 (KD = 0.4 ± 0.1 nM) and BA.2 spike trimers (roughly 0.3 nM) extra tightly than the WT (KD = 0.5 ± 0.3 nM). In pseudovirus neutralization assays, JMB2002 has equal inhibition efficacy in blocking the Omicron pseudovirus an infection of human ACE2-expressing cells. The half-maximal inhibition focus (IC50) was 0.2 μg/mL.
Nonetheless, the L452R mutation within the Delta and BA.4/5 variants is situated on the binding epitope place of JMB2002. This L452R mutation would block the Y102 binding of the heavy chain of JMB2002 Fab. Newly emerged BA.4 and BA.5 carrying L452R mutation and BA.2.12.1 subtype harboring L452Q mutation could evade JMB2002 Fab binding. The immune escape capability of Omicron BA.4/BA.5 is about 20-fold greater than WT pressure, and even threefold greater than BA.1 and BA.23,4 (Fig. 1c). This has raised concern about reinfection and breakthrough an infection among the many recovered or vaccinated inhabitants. Surprisingly, the recurring L452R mutation has been recognized earlier in some BA.1 and recombinant variants (“Deltacron”-like Variants).5 These numerous epitopes from repeatedly evolving variants counsel obligatory cocktail antibody and vaccine antigen regimens to cowl the altered epitopes.
In abstract, the findings from Yin, Xu, and their colleagues present the structural evaluation of the Omicron BA.1 and BA.2 spike trimers and an efficient anti-Omicron BA.1 and BA.2 antibody (JMB2002) together with its binding mechanism, which revealed the interplay between the Omicron spike trimer and hACE2, and found uncommon RBD-RBD interplay mechanism, and instructed potential mouse origin of Omicron variant. The research present the mechanistic foundation for the therapeutic antibody and vaccine improvement in opposition to SARS-CoV-2 and its repeatedly evolving variants. The composite cocktail antigens or antibodies concentrating on the various variant epitopes could contribute to the vaccine and drug improvement and deal with the challenges of repeatedly evolving Omicron variants.