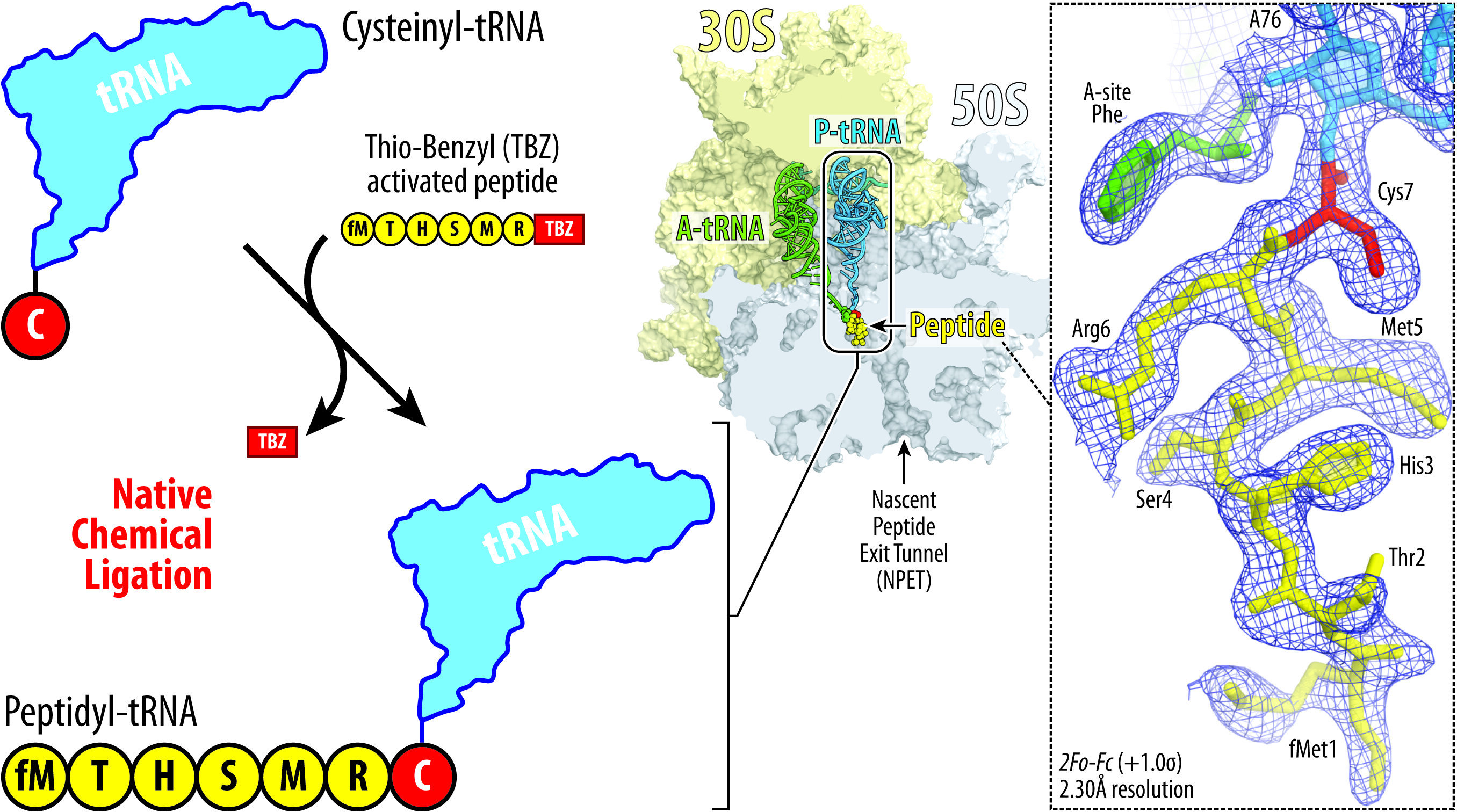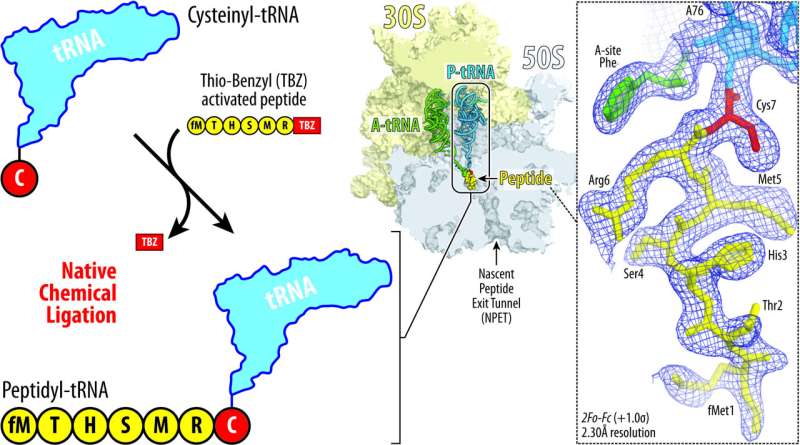
Inside tiny mobile machines known as ribosomes, chains of genetic materials known as messenger RNAs (mRNAs) are matched with the corresponding switch RNAs (tRNAs) to create sequences of amino acids that exit the ribosome as proteins. Unfinished proteins are known as nascent chains, and they’re left connected to the ribosome.
Scientists know that a few of these nascent chains can regulate the exercise of the ribosome and that the nascent chains can typically intrude with antibiotics—a lot of which work by concentrating on bacterial ribosome exercise. Scientists have no idea why this occurs, primarily as a result of it’s laborious to visualise what the ribosome-peptide-drug interactions appear like whereas the unfinished proteins are nonetheless tethered to the ribosome.
Now, scientists on the College of Illinois Chicago are the primary to report a way for steady attachment of peptides to tRNAs, which has allowed them to realize new elementary insights into ribosome perform by figuring out the atomic-level buildings of ribosomes and the shapes that these peptides take contained in the ribosome.
Their technique is newly reported within the journal Nature Chemistry.
“The problem has been to see up shut the construction of the ribosome and the exit tunnel within the presence of the nascent peptides as a result of, in nature, the ribosome could be very fast for us to seize photographs or conduct experiments,” mentioned Yury Polikanov, affiliate professor within the organic sciences division on the School of Liberal Arts and Sciences. “Till the arrival of this new technique, we have basically been blinded from seeing what is occurring within the energetic website of the ribosome at this essential second in time.”
Polikanov and his colleague Egor Syroegin, a Ph.D. candidate in organic sciences at UIC, used a way known as native chemical ligation to fuse customized peptides with the tRNA to yield what is known as a peptidyl-tRNA.
“Acquiring tRNA molecules linked to peptides, just like these contained in the ribosome throughout protein synthesis, has remained a dream of many researchers within the area for nearly 20 years,” Polikanov mentioned. “This has been extraordinarily difficult as a result of there aren’t any enzymes that may immediately connect peptides to a tRNA.”
“The strategy has been used for a very long time in chemistry, but it surely has by no means been utilized on this means. It is mimicking nature, principally, and with our superior imaging expertise, we at the moment are seeing how nature works at a excessive decision,” Syroegin mentioned.
With this new method, Polikanov and Syroegin decided a set of high-resolution buildings of the ribosome carrying peptidyl-tRNAs of varied lengths.
Detailed evaluation of those buildings supplies new and stunning insights into the mechanism of the ribosome’s catalytic middle and answered a number of long-standing elementary questions within the ribosome area, Polikanov mentioned.
“We noticed that relying on the sequence, totally different peptides can kind totally different shapes or folds inside the ribosomal tunnel, and we will synthesize totally different peptides of various sequences after which comply with their form very exactly, due to the excessive decision of our buildings,” Syroegin mentioned. “So now, we will very confidently say that ‘these peptides, of this sequence, have this form’ or ‘one other peptide has one other form.’ That is necessary as a result of nascent peptide folding determines whether or not medicine would arrest ribosome or not.”
“This technique opens numerous avenues for structural and practical research aimed toward understanding the mechanisms of ribosome functioning, in addition to sequence-specific ribosome stalling induced by sure antibiotics,” Polikanov mentioned.
Polikanov and Syroegin are co-authors of the paper “Insights into the ribosome perform from the buildings of non-arrested ribosome nascent chain complexes,” together with Elena Aleksandrova, analysis specialist within the organic sciences division at UIC.
Extra info:
Egor A. Syroegin et al, Insights into the ribosome perform from the buildings of non-arrested ribosome–nascent chain complexes, Nature Chemistry (2022). DOI: 10.1038/s41557-022-01073-1
Supplied by
College of Illinois at Chicago
Quotation:
Staff develops a brand new technique for finding out atomic-level ribosome perform (2022, November 1)
retrieved 1 November 2022
from https://phys.org/information/2022-11-team-method-atomic-level-ribosome-function.html
This doc is topic to copyright. Aside from any truthful dealing for the aim of personal research or analysis, no
half could also be reproduced with out the written permission. The content material is offered for info functions solely.