On this knowledge descriptor, we current the datasets underlying this research, that are saved on the DRYAD knowledge repository, dataset 1–37,8, and examples from every dataset to facilitate preliminary viewing and downloading, Pattern dataset19.
Dataset 1: Volumetric and 3D-spatial OPT assessments of BCM distribution from pancreatic compartments (splenic lobe (SL), duodenal lobe (DL), gastric lobe (GL)) in a SHD and MLD diabetic mice, 1-, 2- and 3-weeks submit injection compared to automobile controls (SHDvCtrl, MLDvCtrl) and untreated controls (Ctrl) with corresponding blood glucose ranges and physique weights (Desk S1)7.
Dataset 2: OPT evaluation of BCM and GLUT2 3D expression intensities in pancreatic splenic lobes from SHD induced hyperglycaemia mice, wherein glycemia was restored by islet transplantation (SHD + Tx). Pancreata have been collected 28 days submit administration of STZ and in comparison with automobile management and SHD constructive management with corresponding blood glucose ranges and physique weights (Desk S2)8.
Dataset 3: Excessive-resolution assessments of islet morphology utilizing LSFM from consultant samples from dataset 18.
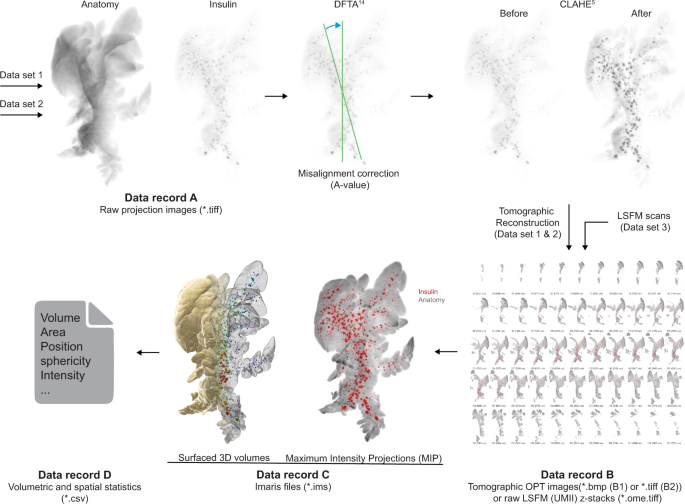
Every dataset is subdivided into knowledge data, based mostly on the picture processing pipeline (see Fig. 3). The offered uncooked projection views (knowledge document A, datasets 1 & 2, knowledge quotation 1 & 5) have been generated by an in home construct close to infrared -OPT scanner16 as *.tiff information. For knowledge document B, tomographic 2D picture datasets have been processed and reconstructed into tomographic sections (datasets 1 & 2, knowledge quotation 2 and 6). Information document B additional consists of unprocessed LSFM generated sections (Dataset 3, knowledge quotation 9). For knowledge document C, Z-sections from OPT and LSFM imaging have been reworked into Imaris (*.ims) information for assessments of spatial and quantitative options of BCM distribution (dataset 1,2 & 3, knowledge quotation 3, 7 & 10). The ensuing quantitative knowledge have been extracted from Imaris as Excel sheets (knowledge document D, knowledge quotation 4 & 8) comprising numerical knowledge on islets volumes, staining intensities, and islet sphericity, along with knowledge on the pancreatic lobular anatomy. Collectively, the introduced datasets could facilitate the planning, execution, and analysis of a variety of analysis undertakings pertaining to STZ-induced diabetes in rodents.
Information processing pipeline. Information document A from dataset 1 (Information document A, Information quotation 1) and a pair of (Information document A, Information quotation 5) have been processed utilizing a set of in-house developed post-scanning computational scripts, DFTA (uniform alignment values) and CLAHE (equalizing the distinction of the insulin labelled islets) previous to reconstruction into tomographic photos (picture processing bundle “DSPOPT”, together with DFTA (“A-value” tuning) and CLAHE may be discovered: https://github.com/ARDISDataset/DSPOPT). Information document B additionally embody LSFM z-stacks from dataset 3 (Information quotation 2, 6 and 9). The tomographic photos transformed to Imaris native.ims information have been analysed (Information document C, 3, 7 and 10). Volumetric and spatial statistics was extracted in Imaris (Information document D, Information quotation 4 and eight).
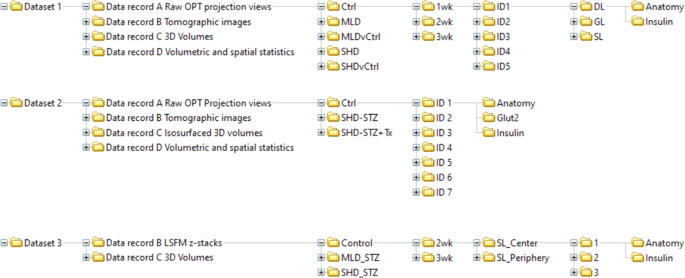
A schematic picture of the organised knowledge tree is displayed in Fig. 4. The info data describe the uncooked, processed (tomographic reconstructions), finish level picture datasets and quantitative/spatial knowledge of the total BCM distribution in STZ-induced diabetic mice (SHD, MLD, SHD + Tx) and their wholesome (C57BL/6) controls at 1-, 2- and 3-weeks post-administration, in addition to automobile controls (SHDvCtrl and MLDvCtrl). Observe, the MLD group on the 1-week time level didn’t have any diabetic animals however have been nonetheless included within the dataset.
Schematic illustration of knowledge folder tree. Schematic illustration depicting the sub-organization of the datasets integrated within the knowledge descriptor together with knowledge data A-D, remedy teams, time factors submit STZ administration, pattern IDs, imaging channels and scan location (LSFM).
Information document A
Uncooked projection datasets generated by OPT may be present in ‘Information document A Uncooked OPT projection views’ (Information document A Uncooked OPT projection views.zip, Information quotation 1 and 5). Every particular person scan is supported by a log file in *.txt format together with scanning parameters akin to publicity instances or rotation steps. The person picture information are titled to point experimental group, age post-administration, animal ID, pancreatic lobe (splenic (SL), duodenal (DL) or gastric (GL)), channel (Insulin or Anatomy or for dataset 2 GLUT2) and step rotation quantity (1 step = 0.9 levels of rotation) of projection picture, e.g., ‘SHD_2wk_ID3_SL_Insulin_0398.tif‘.
Information document B
Information generated by tomographic reconstruction of OPT processed knowledge (Axis of rotation, DFTA and CLAHE) may be present in ‘Information document B Tomographic photos’ (Information document B Tomographic photos.zip, Information quotation 2 and 6), in addition to uncooked generated LSFM z-sections as ‘Information document B LSFM z-stacks’ (Information document B LSFM z-stacks.zip, Information quotation 9). The person picture sections of dataset 1 and a pair of are annotated to point experimental group, age post-administration, animal ID, pancreatic lobe (SL, DL, GL respectively), channel (Insulin or Anatomy, or GLUT2 for dataset 2) and sequential z-stack quantity, e.g., ‘Ctrl_1wk_ID4_DL_Anatomy_0554.bmp’. Every LSFM part in document B in dataset 3 signifies experimental group, age post-administration, location of scanned quantity (periphery or centre of the pancreatic splenic lobe), scan ID, channel (Insulin or Anatomy) and z-stack quantity, e.g., ‘MLD_STZ_2wk_SL_Center_2_Insulin_Z0003.ome.tif.
Information document C
Reconstructed knowledge transformed to *.ims format with 3D iso-surfaced volumes may be present in ‘Information record_C_Isosurfaced quantity information’ (Information record_C_ Isosurfaced quantity information.zip, Information quotation 3 and seven) and for Dataset 3 Quantity information (Information quotation 10). The information include (for datasets 1 and a pair of) iso-surfaces of islets of Langerhans based mostly on the Insulin channel and of the lobular anatomy based mostly on the anatomy channel, and 3D-volumes of the above-mentioned buildings (dataset 3). They’re annotated to point experimental group, age post-administration, animal ID and pancreatic lobe (splenic (SL), duodenal (DL) or gastric (GL)), e.g., ‘Ctrl_3w_ID4_SL.ims.
Information document D
Ensuing quantitative knowledge of processed 3D OPT volumes have been retrieved from Imaris and may be present in ‘Information document D volumetric and spatial statistics’ (Information document D volumetric and spatial statistics.zip, Information quotation 4 and eight). Reconstructed OPT scans produce isotropic voxels, which permits for dependable quantification, whereas LSFM scans typically have a distortion within the z-axis, as do the Ultramicroscope II used on this research because of the era of the sunshine sheet being two cones overlapping into one another somewhat than two parallel traces. Subsequently, quantitative knowledge is given for OPT knowledge solely. The OPT based mostly Excel sheets are uncooked extracted numerical info. File titles point out experimental group, age post-administration, animal ID, pancreatic lobe (splenic (SL), duodenal (DL) or gastric (GL)) and channel info, e.g., ‘Ctrl_3w_ID4_DL_anatomy.csv’. The csv information are subdivided into multiples information, every displaying a unique parameter for the person islets:
-
Quantity
-
Space
-
Place
-
Sphericity
-
Depth (centre, min, max, imply, median, sum)
-
Dimensional (x,y,z) diameter
-
Centre of homogenous mass
-
Distance to picture border
-
Ellipsoid axis
-
Ellipticity (oblate/prolate)
-
Variety of triangles
-
Variety of vertices
-
Variety of voxels
Supplementary Tables 3 and 4 show meta knowledge of every pattern to help knowledge data and point out every samples provenance and experimental manipulations carried out in addition to the ensuing knowledge outputs and which archived data they kind.