Final Up to date: November 4, 2022
Introduction to Nucleic Acids
The metabolic necessities for the nucleotides and their cognate bases may be met by each dietary consumption or synthesis de novo from low molecular weight precursors. Certainly, the power to salvage nucleotides from sources throughout the physique alleviates any vital dietary requirement for nucleotides, thus the purine and pyrimidine bases should not required within the weight loss program. The salvage pathways are a serious supply of nucleotides for synthesis of DNA, RNA and enzyme co-factors.
Inside the physique the main website of de novo nucleotide synthesis, for the replenishment and upkeep of intracellular swimming pools, is the liver. Following their synthesis within the liver the nucleotides are dephosphorylated and partly phosphorolytically cleaved into nucleobases and ribose-1-phosphate for transport to the blood after which subsequent uptake by cells of the opposite organs.
Extracellular hydrolysis of ingested nucleic acids happens by way of the concerted actions of endonucleases, phosphodiesterases, and nucleoside phosphorylases. Endonucleases degrade DNA and RNA at inside websites resulting in the manufacturing of oligonucleotides. Oligonucleotides are additional digested by phosphodiesterases that act from the ends inward yielding free nucleosides. The bases are hydrolyzed from nucleosides by the motion of phosphorylases that yield ribose-1-phosphate and free nucleobases. If the nucleosides and/or bases should not re-utilized the purine bases are additional degraded to uric acid and the pyrimidines to β-aminoiosobutyrate, NH3 and CO2.
Activation of Ribose-5-Phosphate
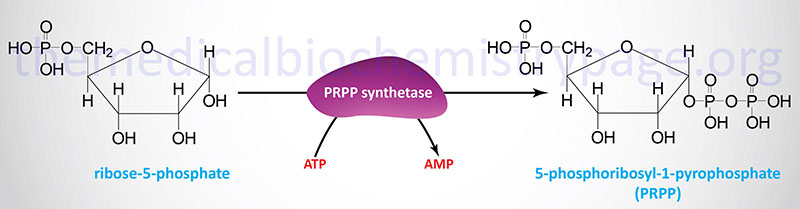
Each the salvage and de novo synthesis pathways of purine and pyrimidine biosynthesis result in manufacturing of nucleoside-5′-phosphates by way of the utilization of an activated sugar intermediate and a category of enzymes referred to as phosphoribosyltransferases. The activated sugar used is 5-phosphoribosyl-1-pyrophosphate, PRPP. PRPP is generated by the motion of PRPP synthetase (additionally referred to as ribose-phosphate pyrophosphokinase 1) and requires vitality within the type of ATP.
At the least three totally different enzymes with PRPP synthetase exercise have been recognized that are encoded by three distinct genes. These genes are recognized as PRPS1, PRPS2, and PRPS1L1 (PRPS1-like 1). The PRPS1 and PRPS2 genes are each positioned on the X chromosome, PRPS1 is on the q arm (Xq22.3) and PRPS2 is on the p arm (Xp22.2). The PRPS1 gene consists of seven exons that generate two alternatively spliced mRNAs encoding isoform 1 (318 amino acids) and isoform 2 (114 amino acids). The PRPS2 gene can be composed of seven exons that generate two alternatively spliced mRNAs encoding isoform 1 (321 amino acids) and isoform 2 (318 amino acids).
The PRPS1L1 gene is an intronless gene positioned on chromosome 7p21.1 that encodes a protein of 318 amino acids. The PRPS1L1 gene is expressed completely within the testes and translation of the ensuing mRNA begins at a non-AUG codon (ACG). Though ACG usually codes for threonine, within the PRPS1L1 mRNA this different begin codon directs the initiator methionine for the encoded protein.
PRPP Synthetase Mutations
Some mutations within the PRPS1 gene are related to PRPP synthetase tremendous exercise. As a result of the PRPS1 gene is on the X chromosome the illness is classed as an X-linked recessive dysfunction. Most affected people expertise a gentle type of the dysfunction that manifests in late adolescence or early maturity. Signs of the gentle type of the dysfunction are primarily associated to extra uric acid manufacturing inflicting uric acid crystalluria and urinary stones, adopted by the event of gouty arthritis. Untreated the dysfunction will end in renal failure from uric acid crystal deposition. The extra extreme kind manifests in infancy or early childhood with signs that embody hypotonia, ataxia, sensorineural listening to loss, developmental delay, and mental impairment,
On the different finish of the spectrum are mutations within the PRPS1 gene that end in extreme or average lack of operate of PRPP synthetase. The dysfunction related to extreme lack of operate known as Arts syndrome. Arts syndrome is related to profound sensorineural listening to loss, hypotonia, ataxia, developmental delay, and mental incapacity predominantly in males. Manifesting females expertise a lot milder signs. In early childhood affected males will develop imaginative and prescient loss, peripheral neuropathy, and may have recurrent infections. On account of the infections and the opposite issues of Arts syndrome, affected males typically don’t survive previous early childhood.
The average lack of PRPP synthetase operate leads to the dysfunction referred to as Charcot-Marie-Tooth illness X-linked recessive sort 5 (CMTX5). CMTX5 is also referred to as Rosenberg-Chutorian syndrome. CMTX5 just isn’t actually a classical type of CMT illness and most investigators really feel the designation is inappropriate for this type of illness which is related to peripheral nerve issues, deafness, and imaginative and prescient loss.
Purine Nucleotide Biosynthesis
Synthesis of Inosine Monophosphate: IMP
The foremost website of purine synthesis is within the liver. Synthesis of the purine nucleotides begins with PRPP and results in the primary totally fashioned nucleotide, inosine 5′-monophosphate (IMP). This pathway is diagrammed under. The purine base with out the connected ribose moiety is hypoxanthine. The purine base is constructed upon the ribose by a number of amidotransferase and transformylation reactions. The synthesis of IMP requires 5 moles of ATP, two moles of glutamine, one mole of glycine, one mole of CO2, one mole of aspartate and two moles of formate. The formyl moieties are carried on tetrahydrofolate (THF) within the type of N10-formyl-THF (10-formyltetrahydrofolate).
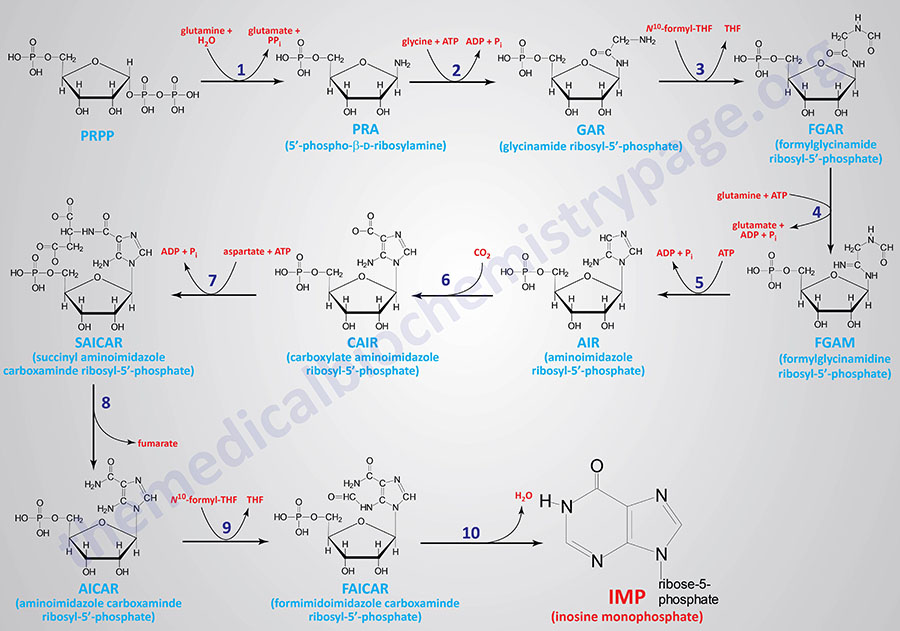
1. glutamine phosphoribosylpyrophosphate amidotransferase (GPAT exercise of the PPAT gene)
2. glycinamide ribonucleotide synthetase (GARS exercise of the GART gene)
3. glycinamide ribonucleotide formyltransferase (GART exercise of the GART gene)
4. phosphoribosylformylglycinamide synthase (PFAS exercise of the PFAS gene)
5. aminoimidazole ribonucleotide synthetase (AIRS exercise of the GART gene)
6. aminoimidazole ribonucleotide carboxylase (AIRC exercise of the PAICS gene)
7. succinylaminoimidazolecarboxamide ribonucleotide synthetase (SAICAR exercise of the PAICS gene)
8. adenylosuccinate lyase (ADSL exercise of the ADSL gene)
9. 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase (AICARFT exercise of the ATIC gene)
10. IMP cyclohydrolase (IMPCH exercise of the ATIC gene)
The primary response (indicated by 1 within the Determine under) of purine synthesis is catalyzed by an enzyme referred to as glutamine phosphoribosylpyrophosphate amidotransferase that’s encoded by the PPAT gene (phosphoribosylpyrophosphate amidotransferase: PRPP amidotransferase). This enzyme is usually referred to easily as amidotransferase. The PPAT gene is positioned on chromosome 4q12 which consists of 11 exons that encode a 517 amino acid protein.
The actions that catalyze reactions 2, 3, and 5 are all contained in a single tri-functional enzyme encoded by the GART gene (phosphoribosyl-glycinamide formyltransferase, phosphoribosyl-glycinamide synthetase, phosphoribosyl-aminoimidazole synthetase). The GART gene is positioned on chromosome 21q22.11 and consists of 24 exons that generate 4 alternatively spliced mRNAs. Three of the mRNAs from the GART gene all encode the identical 1010 amino acid protein.
Response 4 of purine synthesis is catalyzed by phosphoribosyl-formylglycinamide synthase which is encoded by the PFAS gene. The PFAS gene is positioned on chromosome 17p13.1 and consists of 30 exons that encode a protein of 1338 amino acids.
Reactions 6 and 7 are catalyzed by a bi-functional enzyme encoded by the PAICS gene (phosphoribosyl-aminoimidazole carboxylase, phosphoribosyl-aminoimidazole succinocarboxamide synthetase). The PAICS gene is positioned on chromosome 4q12 carefully related to the PPAT gene whose encoded enzyme catalyzes step one of purine synthesis. The PAICS gene consists of 21 exons that generate six alternatively spliced mRNAs that collectively encode 5 totally different protein isoforms.
Expression of each the PPAT and PAICS genes is coordinately regulated.
Response 8 of purine synthesis is catalyzed by adenylosuccinate lyase. Adenylosuccinate lyase is encoded by the ADSL gene. The ADSL gene is positioned on chromosome 22q13.1 and consists of 14 exons that generate seven alternatively spliced mRNAs, every of which encode a definite protein isoform.
Mutations within the ADSL gene are related to adenylosuccinate lyase deficiency (ADSLD). a dysfunction ensuing from the buildup of poisonous intermediates, succinyladenosine and succinylaminoimidazole carboxamide riboside. Three scientific phenotypes of ADSLD have been outlined with probably the most extreme kind being a deadly neonatal encephalopathy. Prognosis of ADSLD is made by measurement of succinyladenosine within the urine.
The final two reactions (9 and 10) are catalyzed by a bi-functional enzyme encoded by the ATIC gene (5-aminoimidazole-4-carboxyamide ribonucleotide formyltransferase, IMP cyclohydrolase). The ATIC gene is positioned on chromosome 2q35 and consists of 20 exons that encode a protein of 592 amino acids.
Synthesis of AMP and GMP from IMP
IMP represents a department level for purine biosynthesis, as a result of it may be transformed into both AMP or GMP by way of two distinct response pathways. The pathway resulting in AMP requires vitality within the type of GTP; that resulting in GMP requires vitality within the type of ATP. The utilization of GTP within the pathway to AMP synthesis permits the cell to manage the proportions of AMP and GMP to close equivalence. The buildup of extra GTP will result in accelerated AMP synthesis from IMP as an alternative, on the expense of GMP synthesis. Conversely, for the reason that conversion of IMP to GMP requires ATP, the buildup of extra ATP results in accelerated synthesis of GMP over that of AMP.
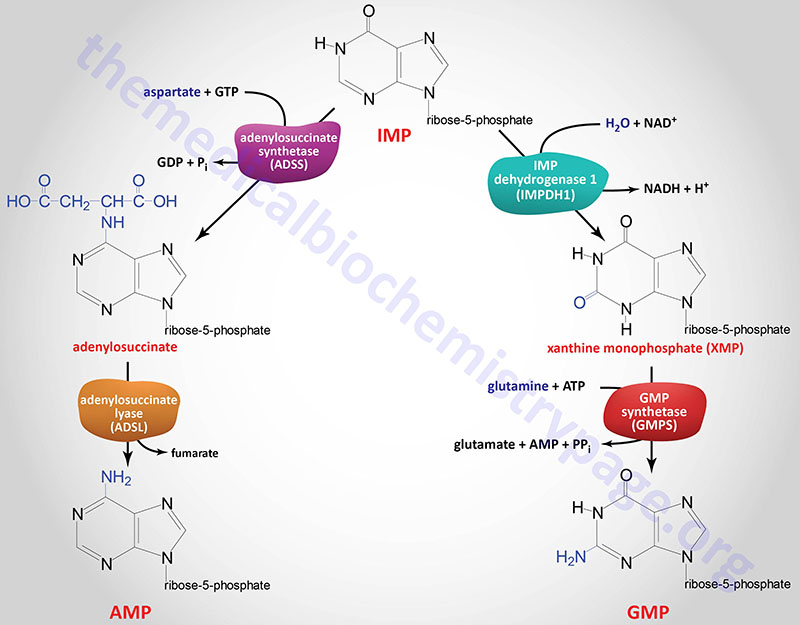
The 2 enzymes within the IMP to AMP pathway are adenylosuccinate synthetase and adenylosuccinate lyase. People specific two adenylosuccinate synthetase genes, ADSS1 and ADSS2.
The ADSS1 gene is positioned on chromosome 14q32.33 and consists of 16 exons that generate three mRNAs by way of different splicing and different promoter utilization. All three mRNAs encode totally different protein isoforms.
The ADSS2 gene is positioned on chromosome 1q44 and consists of 15 that generate two alternatively spliced mRNAs, each of which encode distinct protein isoforms.
The adenylosuccinate lyase on this pathway is identical enzyme that catalyzes response 8 of de novo purine biosynthesis as described above.
The 2 enzymes within the IMP to GMP pathway are inosine-5′-monophosphate dehydrogenase (IMP dehydrogenase: IMPDH) and GMP synthetase. People specific two IMPDH genes recognized as IMPDH1 and IMPDH2.
The IMPDH1 gene is positioned on chromosome 7q32.1 and consists of 18 exons that generate eight alternatively spliced mRNAs encoding eight protein isoforms.
The IMDPH2 gene is positioned on chromosome 3p21.31 and consists of 15 exons that generate 4 alternatively spliced mRNAs, every of which encode a definite protein isoform.
Expression of IMPDH1 predominates within the retina, spleen, and resting peripheral blood mononuclear cells however just like the IMPDH2 gene can be expressed in most tissues at various ranges. Whatever the tissue, IMPDH1 is expressed constitutively at low ranges. Expression of IMPDH2 is enhanced throughout proliferation and transformation.
GMP synthetase is encoded by the GMPS gene. The GMPS gene is positioned on chromosome 3q25.31 and consists of 19 exons that encode a 693 amino acid protein.
Regulation of Purine Nucleotide Synthesis
The important charge limiting steps in purine biosynthesis happen on the first two steps of the pathway. The synthesis of PRPP by PRPP synthetase is feed-back inhibited by purine-5′-nucleotides (predominantly AMP and GMP). Combinatorial results of these two nucleotides are best, e.g., inhibition is maximal when the right focus of each adenine and guanine nucleotides is achieved.
The amidotransferase response catalyzed by PRPP amidotransferase can be feed-back inhibited allosterically by binding ATP, ADP and AMP at one inhibitory website and GTP, GDP and GMP at one other. Conversely the exercise of the enzyme is stimulated by PRPP.
Moreover, purine biosynthesis is regulated within the department pathways from IMP to AMP and GMP. The buildup of extra ATP results in accelerated synthesis of GMP, and extra GTP results in accelerated synthesis of AMP.
Catabolism of Purine Nucleotides
Catabolism of the purine nucleotides (each ribonucleotides and deoxyribonucleotides) leads in the end to the manufacturing of uric acid which is insoluble and is excreted within the urine. Uric acid excretion and reabsorption happens throughout the proximal tubules of the kidney. Elevation in uric acid ranges may end up in precipitation of urate crystals with monosodium urate crystals being probably the most generally occurring within the synovial fluids of the joints.
The catabolism of purine nucleotides entails deamination response, phosphate elimination from the nucleoside monophosphates, phosphorylytic elimination of the ribose yielding ribose-1-phosphate, and eventually oxidation of the nucleobases to uric acid.
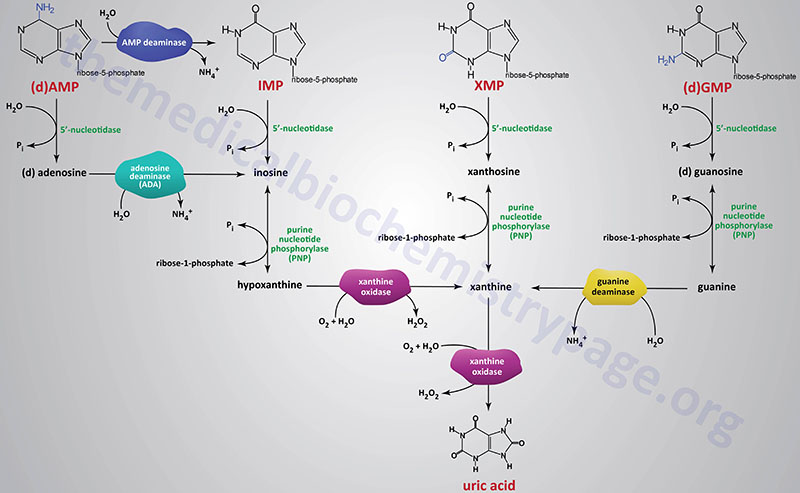
Dephosphorylation of nucleoside monophosphates is catalyzed by 5′-nucleotidases. People specific seven 5′-nucleotidase genes with 5 encoding cytosolic enzymes, one encoding a mitochondrially localized enzyme, and one gene encoding an extracellular enzyme that’s tethered to the plasma membrane by way of a GPI linkage.
Catabolism of AMP and dAMP can happen by conversion first to IMP by way of the motion of AMP deaminase or following 5′-nucleotidase motion producing adenosine or deoxyadenosine. On this latter pathway the nitrogen is faraway from adenosine producing inosine by the essential enzyme, adenosine deaminase, ADA.
People specific three AMP deaminase genes: AMPD1, AMPD2, and AMPD3.
The AMPD1 gene is positioned on chromosome 1p13.2 and consists of 16 exons that generate two alternatively spliced mRNAs encoding isoform 1 (747 amino acids) and isoform 2 (776 amino acids). The AMPD1 encoded enzymes are primarily liable for skeletal muscle deamination of AMP. Mutations within the AMPD1 gene characterize the commonest reason for metabolic myopathy.
The AMPD2 gene is positioned on chromosome 1p13.3 and consists of 20 exons that generate 5 alternatively spliced mRNAs that collectively encode 4 distinct protein isoforms. The AMPD2 encoded enzymes are primarily liable for liver deamination of AMP.
The AMPD3 gene is positioned on chromosome 11p15.4 and consists of 19 exons that generate 5 alternatively spliced mRNAs that collectively encode 4 distinct protein isoforms. The AMPD3 encoded enzymes are primarily liable for erythrocyte deamination of AMP.
The ADA gene is positioned on chromosome 20q13.12 and consists of 12 exons that generate three alternatively spliced mRNAs, every of which encode a definite protein isoform. The best ranges of ADA expression are noticed within the duodenum of the small intestines. Lack of ADA exercise leads to the doubtless deadly dysfunction, extreme mixed immunodeficiency, SCID.
The ribose is faraway from the nucleotides by purine nucleoside phosphorylase (PNP) yielding the nucleobases, hypoxanthine, xanthine, and guanine. Purine nucleotide phosphorylase is encoded by the PNP gene. The PNP gene is positioned on chromosome 14q11.2 and consists of 6 exons that encode a 289 amino acid protein.
The nitrogen is faraway from guanine by guanine deaminase yielding xanthine. Guanine deaminase is encoded by the GDA gene. The GDA gene is positioned on chromosome 9q21.13 and consists of 27 exons that generate seven alternatively spliced mRNAs that collectively encode 5 distinct protein isoforms.
Hypoxanthine and xanthine are then transformed to the terminal product of purine catabolism, uric acid, by the enzyme xanthine oxidase. The enzymatic exercise referred to as xanthine oxidase is the time period used for the modified from of the enzyme xanthine dehydrogenase which is a molybdenum-dependent hydroxylase that capabilities as a homodimer. The conversion xanthine dehydrogenase to xanthine oxidase outcomes from reversible sulfhydryl oxidation in addition to from irreversible proteolytic motion.
Xanthine dehydrogenase is encoded by the XDH gene. The XDH gene is positioned on chromosome 2p23.1 and consists of 37 exons that generate a 1337 amino acid protein.
Salvage of Purine Nucleotides
The re-synthesis of nucleotides from the purine bases and purine nucleosides takes place in a collection of steps often known as the salvage pathways. The free purine bases, adenine, guanine, and hypoxanthine, may be reconverted to their corresponding nucleotides by phosphoribosylation the place PRPP, like within the de novo synthesis pathway, serves because the activated type of ribose-5′-phosphate.
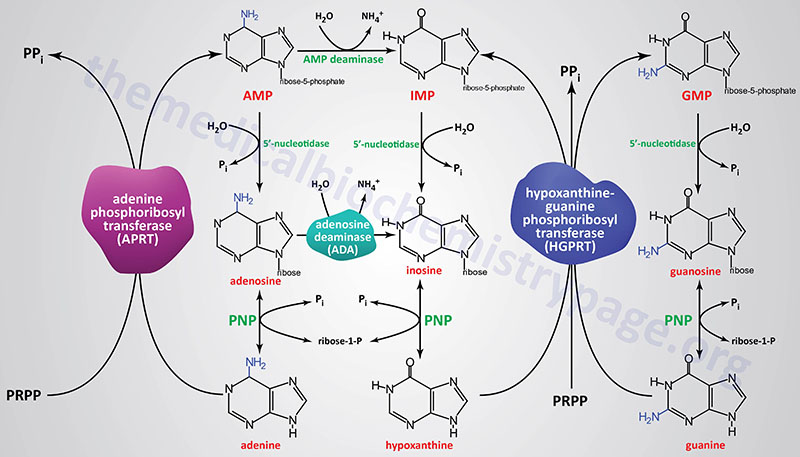
Most all cells perform purine nucleotide salvage, with the notable exception being lymphocytes through which the actions of the purine salvage pathways are lowered in comparison with different cells which makes lymphocytes significantly depending on the pathways of de novo purine synthesis for proliferation. This dependence of lymphocyte proliferation on de novo purine synthesis makes them significantly inclined to inhibition of enzymes corresponding to IMP dehydrogenase. Mycophenolic acid, an inhibitor of each the IMPDH1 and IMPDH2 encoded enzymes is usually used as an immunosuppressant in sufferers following kidney transplantation.
Two key transferase enzymes are concerned within the salvage of purines: adenosine phosphoribosyltransferase (APRT), which catalyzes the next response:
adenine + PRPP ↔ AMP + PPi
and hypoxanthine-guanine phosphoribosyltransferase (HGPRT), which catalyzes the next reactions:
hypoxanthine + PRPP ↔ IMP + PPi
guanine + PRPP ↔ GMP + PPi
Adenosine phosphoribosyltransferase is encoded by the APRT gene. The APRT gene is positioned on chromosome 16q24.3 and consists of 5 exons that generate two alternatively spliced mRNAs encoding isoform a (180 amino acids) and isoform b (134 amino acids).
Hypoxanthine-guanine phosphoribosyltransferase is encoded by the HPRT1 gene. The HPRT1 gene is positioned on the X chromosome (Xq26.2–q26.3) and consists of 9 exons that encode a 218 amino acid protein.
A critically vital enzyme of purine salvage in quickly dividing cells is adenosine deaminase (ADA) which catalyzes the deamination of adenosine to inosine. Deficiency in ADA leads to the dysfunction referred to as extreme mixed immunodeficiency, SCID (and briefly outlined under).
Extra enzymes are utilized within the salvage of the deoxynucleotides. These extra enzymes have overlapping specificities and their actions management the speed limiting phosphorylation of deoxynucleosides. These extra enzymes are deoxycytidine kinase, deoxyguanosine kinase, and the 2 types of thymidine kinase, one that’s cytosolic (TK1) and one that’s mitochondrial (TK2).
Deoxycytidine kinase is encoded by the DCK gene which is positioned on chromosome 4q13.3 and consists of 8 exons that generate seven alternatively spliced mRNAs that collectively encode 5 distinct protein isoforms. Deoxycytidine kinase can phosphorylate deoxyadenine, deoxycytidine, and deoxyguanosine.
Deoxyguanosine kinase is a mitochondrial enzyme that’s encoded by the DGUOK gene. The DGUOK gene is positioned on chromosome 2p13.1 and consists of 8 exons that generate seven alternatively spliced mRNAs, that collectively encode 5 distinct protein isoforms. Deoxyguanosine kinase can phosphorylate deoxyadenine, deoxycytidine, and deoxyguanosine within the mitochondria.
The TK1 gene is positioned on chromosome 17q25.3 and consists of 6 exons that generate three alternatively spliced mRNAs, every of which encode a definite protein isoform. The TK1 encoded proteins phosphorylate thymidine producing dTMP. Expression of the TK1 gene is regulated in a cyclic method with peak expression throughout S-phase (DNA synthesis). The product of the TK1 response, dTMP, is in the end transformed to dTTP by way of the actions of thymidylate kinase after which nucleotide diphosphate kinases. The resultant dTTP serves as a feed-back inhibitor of the thymidine kinase, thus regulating the mobile ranges of dTTP.
The TK2 gene is positioned on chromosome 16q21 and consists of 11 exons that generate seven alternatively spliced mRNAs, every of which encode a definite protein isoform. The TK2 encoded proteins are liable for mitochondrial DNA replication by phosphorylation of thymidine, deoxycytidine, and deoxyuridine.
Purine Nucleotide Cycle
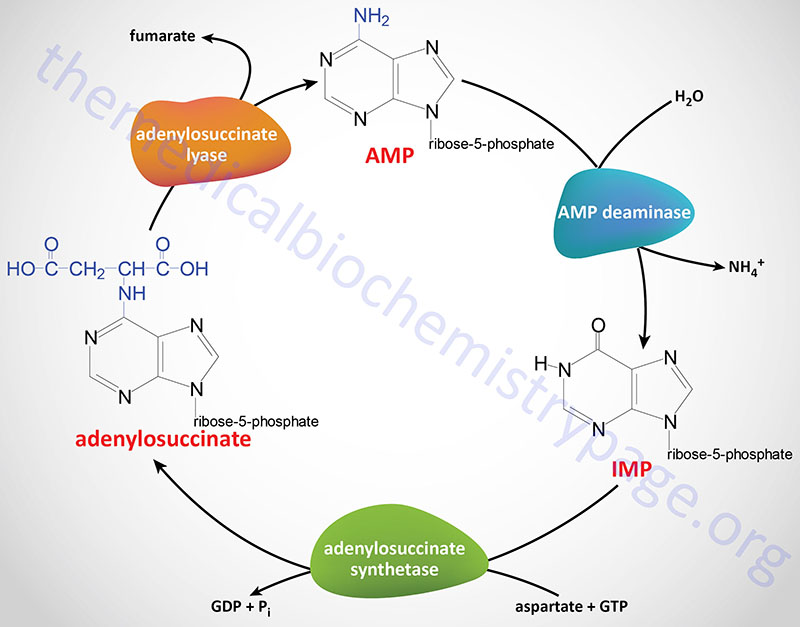
The synthesis of AMP from IMP and the salvage of IMP by way of AMP catabolism have the web impact of deaminating aspartate to fumarate. This course of has been termed the purine nucleotide cycle (see diagram under). This cycle is essential in muscle cells. Throughout intense muscle exercise the demand for ATP will increase with a concomitant demand to extend the TCA cycle, to be able to generate extra NADH for the manufacturing of ATP. Nevertheless, skeletal muscle lacks lots of the enzymes of the main anaplerotic reactions such that fumarate, produced by way of the purine nucleotide cycle, in addition to succinate, and malate characterize the first anaplerotic intermediates for enhanced muscle TCA cycle exercise.
Within the kidney the purine nucleotide cycle represents a way to launch ammonia with out will increase in blood ranges.
Medical Significances of Purine Metabolism
Medical issues related to nucleotide metabolism in people are predominantly the results of irregular catabolism of the purines. The scientific penalties of irregular purine metabolism vary from gentle to extreme and even deadly problems. Medical manifestations of irregular purine catabolism come up from the insolubility of the degradation byproduct, uric acid.
Gout is a situation that outcomes from the precipitation of urate as monosodium urate (MSU) or calcium pyrophosphate dihydrate (CPPD) crystals within the synovial fluid of the joints, resulting in extreme irritation and arthritis. The inflammatory response is as a result of crystals partaking the caspase-1-activating inflammasome ensuing within the manufacturing of interleukin-1β (IL-1β) and IL-18. Most types of gout are the results of extra purine manufacturing and consequent catabolism or to a partial deficiency within the salvage enzyme, HGPRT. Most types of gout may be handled by administering the antimetabolite: allopurinol. This compound is a structural analog of hypoxanthine that strongly inhibits xanthine oxidase.
Two extreme problems, each fairly properly described, are related to defects in purine metabolism: Lesch-Nyhan syndrome and extreme mixed immunodeficiency illness (SCID). Lesch-Nyhan syndrome is an X-linked recessive dysfunction that outcomes from the lack of a purposeful hypoxanthine-guanine phosphoribosyltransferase which is encoded by the HPRT1 gene (additionally typically referred to as HGPRT). Sufferers who inherit lack of operate mutations within the HPRT1 gene exhibit not solely extreme signs of gout but additionally a extreme malfunction of the nervous system. In probably the most severe circumstances, sufferers resort to self-mutilation. Frequent intervention within the self mutilation habits is to surgically take away the sufferers tooth following acquisition of the grownup tooth. Loss of life normally happens earlier than sufferers attain their twentieth yr.
SCID refers to a gaggle of doubtless deadly problems because of a mixed lack of operate of each T- and B-lymphocytes. There are a minimum of 13 identified and characterised genetic causes of SCID. The commonest (45%) reason for SCID is the X-linked dysfunction ensuing from lack of operate of the widespread gamma (γ) chain of the T-cell receptor and different interleukin (IL) receptors. The second commonest (15%) type of SCID is brought on by defects within the enzyme adenosine deaminase, ADA. That is the enzyme liable for changing adenosine to inosine within the catabolism of the purines. This deficiency selectively results in a destruction of B and T lymphocytes, the cells that mount immune responses. Within the absence of ADA, deoxyadenosine is phosphorylated to yield ranges of dATP which are 50-fold greater than regular. The degrees are particularly excessive in lymphocytes, which have ample quantities of the salvage enzymes, together with nucleoside kinases. Excessive concentrations of dATP inhibit ribonucleotide reductase (see under), thereby stopping different dNTPs from being produced. The web impact is to inhibit DNA synthesis.
Since lymphocytes should be capable of proliferate dramatically in response to antigenic problem, the shortcoming to synthesize DNA critically impairs the immune responses, and the illness The accumulating dATP additionally induces DNA strand breakage in non-dividing lymphocytes by way of a direct activation of a serious protease (caspase 9) concerned in apoptosis (programmed cell loss of life). As well as, S-adenosylhomocysteine hydrolase exercise is markedly inhibited by 2′-deoxyadenosine leading to accumulation of S-adenosylhomocysteine which in flip leads to lowered synthesis of S-adenosylmethionine (AdoMet), a essential substrate in transmethylation reactions. ADA poor SCID is normally deadly in infancy until particular protecting measures are taken. A much less extreme immunodeficiency outcomes when there’s a lack of purine nucleoside phosphorylase (PNP) exercise, one other purine degrading enzyme.
One of many many glycogen storage illnesses von Gierke illness additionally results in extreme uric acid manufacturing. This dysfunction outcomes from a deficiency in glucose 6-phosphatase exercise. The elevated availability of glucose-6-phosphate will increase the speed of flux by way of the pentose phosphate pathway, yielding an elevation within the degree of ribose-5-phosphate and consequently PRPP. The will increase in PRPP then end in extra purine biosynthesis adopted by catabolism to uric acid.
Desk of Problems of Purine Metabolism
| Dysfunction | Defect | Nature of Defect | Feedback |
| Gout | PRPP synthetase | elevated enzyme exercise | hyperuricemia |
| Gout | HGPRTa | enzyme deficiency | hyperuricemia |
| Gout | Glucose-6-phosphatase | enzyme deficiency | hyperuricemia; enzyme is poor in von Gierke illness |
| Lesch-Nyhan syndrome | HGPRT | lack of enzyme | related to weird self-mutilating habits along with hyperuricemia and gout |
| SCID | ADAb | lack of enzyme | ADA poor SCID represents 15% of all circumstances of SCID |
| Immunodeficiency | PNPc | lack of enzyme | decreased T-cells; elevated threat for autoimmune problems; additionally related to spasticity, developmental delay, mental incapacity, and ataxia |
| Renal lithiasis | APRTd | lack of enzyme | related to extra manufacturing and renal excretion of two,8-dihydroxyadenine (DHA) leading to DHA crystal nephropathy |
| Xanthinuria | Xanthine oxidase | lack of enzyme | very low uric acid (hypouricemia) related to excessive ranges of xanthine in blood and urine; can result in renal xanthine stones and kidney failure |
| von Gierke illness | Glucose-6-phosphatase | enzyme deficiency | see above |
- ahypoxanthine-guanine phosphoribosyltransferase
- badenosine deaminase
- cpurine nucleotide phosphorylase
- dadenosine phosphoribosyltransferase
Pyrimidine Nucleotide Biosynthesis
Synthesis of the pyrimidines is much less complicated than that of the purines, for the reason that base is far less complicated. The primary accomplished base is derived from one mole of glutamine, one mole of ATP and one mole of CO2 (which kind carbamoyl phosphate) and one mole of aspartate. An extra mole of glutamine and ATP are required within the conversion of UTP to CTP. The pathway of pyrimidine biosynthesis is diagrammed under. The synthesis of pyrimidines differs in two vital methods from that of purines. First, the ring construction is assembled as a free base, not constructed upon PRPP. PRPP is added to the primary totally fashioned pyrimidine base (orotic acid), forming orotate monophosphate (OMP), which is subsequently decarboxylated to UMP. Second, there isn’t a department within the pyrimidine synthesis pathway.
The carbamoyl phosphate used for pyrimidine nucleotide synthesis is derived from glutamine and bicarbonate, throughout the cytosol, versus the urea cycle carbamoyl phosphate derived from ammonia and bicarbonate within the mitochondrion. The urea cycle response is catalyzed by carbamoyl phosphate synthetase 1 (CPS-1, CPS-I) whereas the pyrimidine nucleotide precursor is synthesized by the CPS-2 (CPS-II) exercise of the tri-functional rate-limiting enzyme of pyrimidine nucleotide biosynthesis.
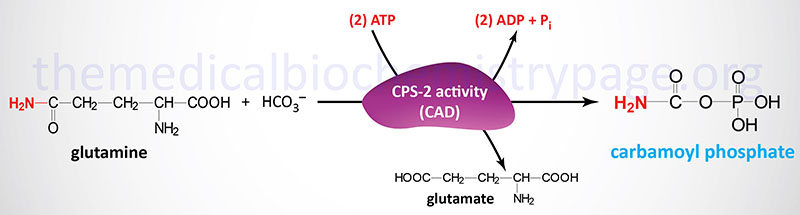
The carbamoyl phosphate that’s produced by this enzyme is then condensed with aspartate within the second step of the response catalyzed by the aspartate transcarbamoylase (ATCase) exercise of the enzyme.
The third step of pyrimidine nucleotide biosynthesis is catalyzed by the dihydroorotase exercise (beforehand known as carbamoyl aspartate dehydratase) of the tri-functional enzyme. The official identify for this tri-functional enzyme is carbamoyl-phosphate synthetase 2, aspartate transcarbamoylase, and dihydroorotase.
Carbamoyl-phosphate synthetase 2, aspartate transcarbamoylase, and dihydroorotase is encoded by the CAD gene. The CAD gene is positioned on chromosome 2p23.3 which consists of 45 exons that generate two alternatively spliced mRNAs, one encoding a protein of 2225 amino acids (isoform 1)and the opposite a protein of 2162 amino acids (isoform 2).
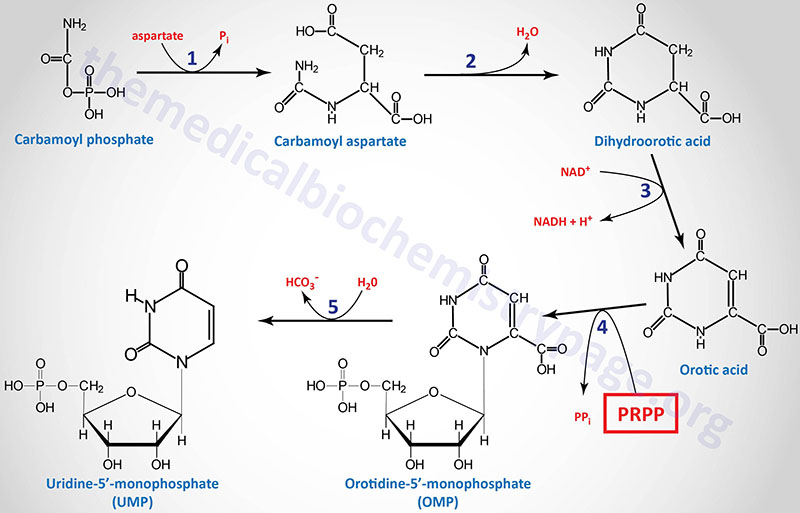
1. aspartate transcarbamoylase (ATCase) exercise of the CAD gene encoded enzyme
2. dihydroorotase (beforehand referred to as carbamoyl aspartate dehydratase) exercise of the CAD gene encoded enzyme
3. dihydroorotate dehydrogenase (DHODH gene exercise)
4. orotate phosphoribosyltransferase exercise of the UMPS gene
5. orotidine-5′-phosphate decarboxylase (OMP decarboxylase) exercise of the UMPS gene
Following the synthesis of dihydroorotic acid by the tri-functional CAD enzyme the compound is oxidized to orotic acid by dihydroorotate dehydrogenase (response 3). Dihydroorotate dehydrogenase is a mitochondrial enzyme tethered to the outer face of the internal mitochondrial membrane.
Dihydroorotate dehydrogenase is encoded by the DHODH gene. The DHODH gene is positioned on chromosome 16q22.2 and consists of 10 exons that encode a protein of 395 amino acids.
The final two reactions (4 and 5), which generate the primary totally fashioned pyrimidine nucleotide (UMP), are catalyzed by the bi-functional homodimeric enzyme recognized as UMP synthetase. This enzyme possesses orotate phosphoribosyltransferase exercise within the N-terminal area and OMP decarboxylase exercise within the C-terminal area. UMP synthetase is encoded by the UMPS gene. The UMPS gene is positioned on chromosome 3q21.2 and consists of seven exons that encode a protein of 480 amino acids.
Following the completion of UMP synthesis this nucleotide is phosphorylated twice to yield UTP (ATP is the phosphate donor). The primary phosphorylation is catalyzed by uridylate kinase and the second by ubiquitous nucleoside diphosphate kinase. The synthesis of CTP happens by way of the amination of UTP by the motion of CTP synthase.
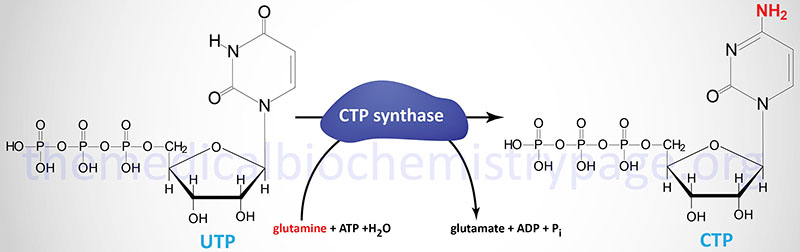
People possess two distinct CTP synthase genes, CTPS1 and CTPS2. The CTPS1 gene is positioned on chromosome 1p34.2 and consists of twenty-two exons that generate two alternatively spliced mRNAs, each of which encode distinct protein isoforms.
The CTPS2 gene is positioned on chromosome Xp22.2 and consists of 25 exons that generate three alternatively spliced mRNAs, every of which encode the identical 586 amino acid protein.
Uridine nucleotides are additionally the precursors for de novo synthesis of the thymine nucleotides (subsequent part). The thymine nucleotides are in flip derived by de novo synthesis from dUMP or by salvage pathways from deoxyuridine or deoxythymidine.
Synthesis of the Thymine Nucleotides
The de novo pathway to dTTP synthesis first requires using dUMP from the metabolism of both UDP or CDP. The dUMP is transformed to dTMP (5-methyl-dUMP) by the motion of thymidylate synthetase. The methyl group (recall that thymine is 5-methyl uracil) is donated by N5,N10-methylene THF. The distinctive property of the motion of thymidylate synthetase is that in the midst of the response the THF is transformed to dihydrofolate (DHF), the one such response yielding DHF from a THF spinoff.
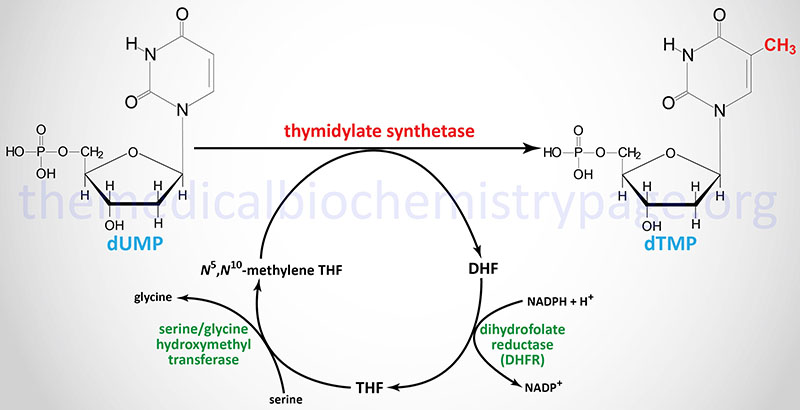
Thymidylate synthetase is encoded by the TYMS gene. The TYMS gene is positioned on chromosome 18p11.32 and consists of 8 exons that generate three alternatively spliced mRNAs, every of which encode distinct protein isoforms. A naturally occurring antisense RNA (NAT) is derived by transcription in the other way to a portion of the three′-end of the TYMS gene. This NAT was initially recognized as rTSα however the gene is now recognized as ENOSF1 (enolase superfamily member 1). Expression of TYMS and the antisense RNA varies inversely throughout development by way of the cell cycle.
To ensure that the thymidylate synthetase response to proceed, THF have to be regenerated from DHF. That is achieved by way of the motion of dihydrofolate reductase which is encoded by the DHFR gene. The DHFR gene is positioned on chromosome 5q14.1 and consists of 6 exons that generate three alternatively spliced mRNAs that encode three distinct isoforms of DHFR.
THF is then transformed to N5,N10-methylene THF by way of the motion of serine hydroxymethyl transferase (encoded by the SHMT1 gene). The essential position of DHFR in thymidine nucleotide biosynthesis makes it an excellent goal for chemotherapeutic brokers (see under).
Salvage of Deoxynucleotides
The salvage pathway to dTTP synthesis entails the thymidine kinase enzymes which might use both thymidine or deoxyuridine as substrate:
thymidine + ATP ↔ TMP + ADP
deoxyuridine + ATP ↔ dUMP + ADP
As indicated above, people specific two thymidine kinase genes, one whose encoded protein is cytosolic (TK1) and the opposite mitochondrial (TK2). The exercise of the TK1 gene is exclusive in that it fluctuates with the cell cycle, rising to peak exercise throughout DNA synthesis (S-phase). Expression of the TK2 gene doesn’t change through the cell cycle.
Medical Relevance of Tetrahydrofolate
Tetrahydrofolate (THF) is regenerated from the dihydrofolate (DHF) product of the thymidylate synthetase response by the motion of dihydrofolate reductase (DHFR), an enzyme that requires NADPH. Cells which are unable to regenerate THF undergo faulty DNA synthesis and eventual loss of life. Because of this, in addition to the truth that dTTP is utilized solely in DNA, it’s therapeutically attainable to focus on quickly proliferating cells over non-proliferating cells by way of the inhibition of thymidylate synthetase or by way of inhibition of DHFR.
The category of molecules used to inhibit thymidylate synthetase are fluorinated pyrimidines. Molecules of this class embody 5-fluorouracil (fluorouracil: 5-FU), 5-fluorodeoxyuridine (floxuridine; FUdR), and 5-fluoro-2′-deoxyuridine monophosphate (FUdR-MP). Fluorouracil inside cells to quite a few metabolic intermediates together with FUdR-MP (additionally written FdUMP), 5-fluorodeoxyuridine triphosphate (FdUTP), and 5-fluorouridine triphosphate (FUTP). It’s FUdR-MP (FdUMP) that inhibits thymidylate synthetase. The opposite main metabolites, FdUTP and FUTP, inhibit DNA synthesis and restore and RNA synthesis, respectively. The various totally different DHFR inhibitors are termed antifolates or antimetabolites and embody methotrexate, aminopterin, trimethoprim, and pyrimethamine. Every of those is a structural analog of folic acid, therefore the time period antifolate.
Regulation of Pyrimidine Biosynthesis
The regulation of pyrimidine synthesis happens primarily at step one which is catalyzed by the trifunctional enzyme encoded by the CAD gene. The ATCase exercise of the enzyme is inhibited by CTP and activated by ATP. The carbamoyl-phosphate synthetase exercise of this complicated is termed carbamoyl-phosphate synthetase 2 (CPS-2), versus CPS-1 which is concerned within the urea cycle. The CAD encoded enzyme is localized to the cytoplasm and the CPS-2 exercise makes use of glutamine because the nitrogen donor for the synthesis of carbamoyl phosphate. CPS-1 of the urea cycle is localized within the mitochondria and makes use of ammonia. The CPS-2 area is activated by ATP and inhibited by UDP, UTP, dUTP, and CTP.
The position of glycine within the regulation of the CAD gene tri-functional enzyme is exerted on the ATCase exercise of the complicated. ATP ranges additionally regulate pyrimidine nucleotide biosynthesis on the degree of PRPP formation. A rise within the degree of PRPP leads to an activation of pyrimidine synthesis.
There’s additionally regulation of OMP decarboxylase exercise of the bi-functional OMP synthase enzyme. The decarboxylase exercise area is competitively inhibited by UMP and, to a lesser diploma, by CMP. Lastly, CTP synthase is feedback-inhibited by CTP and activated by GTP.
Catabolism and Salvage of Pyrimidine Nucleotides
Catabolism of the pyrimidine nucleotides leads in the end to β-alanine (when CMP and UMP are degraded) or β-aminoisobutyrate (when dTMP is degraded) and NH3 and CO2. The β-alanine and β-aminoisobutyrate function -NH2 donors in transamination of α-ketoglutarate to glutamate. A subsequent response converts the merchandise to malonyl-CoA (which may be diverted to fatty acid synthesis) or methylmalonyl-CoA (which is transformed to succinyl-CoA and may be shunted to the TCA cycle).
The salvage of pyrimidine bases has much less scientific significance than that of the purines, owing to the solubility of the by-products of pyrimidine catabolism. Nevertheless, as indicated above, the salvage pathway to thymidine nucleotide synthesis is particularly vital within the preparation for cell division. Uracil may be salvaged to kind UMP by way of the concerted motion of uridine phosphorylase and uridine kinase, as indicated:
uracil + ribose-1-phosphate ↔ uridine + Pi
uridine + ATP ↔ UMP + ADP
Deoxyuridine can be a substrate for uridine phosphorylase. Formation of dTMP, by salvage of thymidine, requires thymine phosphorylase and the beforehand mentioned cytoplasmic thymidine kinase 1 (TK1):
thymine + deoxyribose-1-phosphate ↔ thymidine + Pi
thymidine + ATP → dTMP + ADP
The salvage of deoxycytidine is catalyzed by deoxycytidine kinase:
deoxycytidine + ATP ↔ dCMP + ADP
Deoxyadenosine and deoxyguanosine are additionally substrates for deoxycytidine kinase, though the Okaym for these substrates is far greater than for deoxycytidine.
The foremost operate of the pyrimidine nucleoside kinases is to take care of a mobile stability between the extent of pyrimidine nucleosides and pyrimidine nucleoside monophosphates. Nevertheless, for the reason that total mobile and plasma concentrations of the pyrimidine nucleosides, in addition to these of ribose-1-phosphate, are low, the salvage of pyrimidines by these kinases is comparatively inefficient.
Medical Significances of Pyrimidine Metabolism
As a result of the merchandise of pyrimidine catabolism are soluble, few problems outcome from extra ranges of their synthesis or catabolism. Two inherited problems affecting pyrimidine biosynthesis are the results of deficiencies within the bifunctional enzyme catalyzing the final two steps of UMP synthesis, orotate phosphoribosyltransferase and OMP decarboxylase. These deficiencies end in two sorts of orotic aciduria (sort 1 and kind 2) that trigger retarded progress, and extreme anemia related to hypochromic erythrocytes and megaloblastic bone marrow, each of that are the results of the block to DNA synthesis. Leukopenia can be widespread in orotic acidurias. These problems may be handled with uridine and/or cytidine, which results in elevated UMP manufacturing by way of the motion of nucleoside kinases. The UMP then inhibits the CPS-2 exercise of the CAD encoded enzyme, thus attenuating orotic acid manufacturing.
Desk of Problems of Pyrimidine Metabolism
| Dysfunction | Faulty Enzyme | Feedback |
| Orotic aciduria, sort 1 | because of defects in each the orotate phosphoribosyltransferase and OMP decarboxylase actions of the bifunctional enzyme encoded by the UMPS gene | see above for particulars NOrelated hyperammonemia; regular BUN measurements |
| Orotic aciduria, sort 2 | because of defects within the OMP decarboxylase exercise of the bifunctional enzyme encoded by the UMPS gene | see above for particulars NO related hyperammonemia; regular BUN measurements |
| Orotic aciduria because of OTC deficiency (no hematologic element) |
the urea cycle enzyme ornithine transcarbamylase is poor | elevated mitochondrial carbamoyl phosphate exits and augments cytoplasmic pyrimidine biosynthesis; hepatic encephalopathy; related to hyperammonemia; is NOT related to megaloblastic anemia since no defect in nucleotide metabolism |
| β-aminoisobutyric aciduria | transaminase, impacts urea cycle operate throughout deamination of α-amino acids to α-keto acids | clinically benign; frequent in Orientals |
| drug induced orotic aciduria | OMP decarboxylase exercise of the bifunctional enzyme encoded by the UMPS gene | allopurinol and 6-azauridine therapies trigger orotic acidurias with out a hematologic element; their catabolic by-products inhibit the OMP decarboxylase exercise of the UMPS encoded bifunctional enzyme |
Formation of Deoxyribonucleotides
The everyday cell comprises 5 to10 occasions as a lot RNA (mRNAs, rRNAs and tRNAs) as DNA. Due to this fact, the vast majority of nucleotide biosynthesis has as its goal the manufacturing of rNTPs. Nevertheless, as a result of proliferating cells want to duplicate their genomes, the manufacturing of dNTPs can be essential. This course of begins with the discount of rNDPs, adopted by phosphorylation to yield the dNTPs. The phosphorylation of dNDPs to dNTPs is catalyzed by the identical nucleoside diphosphate kinases that phosphorylates rNDPs to rNTPs, utilizing ATP because the phosphate donor.
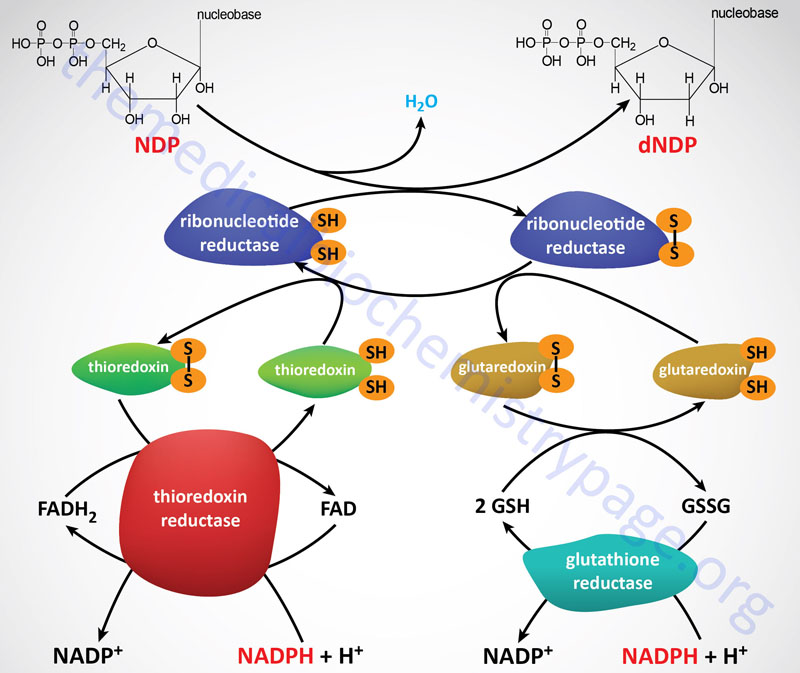
Ribonucleotide reductase (RR) is a heterotetrameric enzyme that comprises redox-active thiol teams for the switch of electrons through the ribonucleotide discount reactions. Practical RR complexes are composed of two catalytic subunits recognized as M1 (massive subunit) and M2 (small subunit). Inside the purposeful RR complicated there are two copies of the M1 subunit and two copies of the M2 subunit. As well as, there’s a regulatory subunit recognized as M2B. The gene encoding the M2B subunit (RRM2B) is inducible by the p53 tumor suppressor.
The M1 protein is encoded by the RRM1 gene. The RRM1 gene is positioned on chromosome 11p15.4 and consists of 20 exons generate 4 alternatively spliced mRNAs, every of which encode a definite protein isoform. The RRM1 gene resides within the area of chromosome 11 that’s deleted in Beckwith-Wiedemann syndrome.
The M2 protein is encoded by the RRM2 gene. The RRM2 gene is positioned on chromosome 2p25.1 and consists of 14 exons that generate two alternatively spliced mRNAs encoding two distinct protein isoforms (isoform 1: 449 amino acids and isoform 2: 389 amino acids).
The M2B protein is encoded by the RRM2B gene. The RRM2B gene is positioned on chromosome 8q22.3 and consists of 9 exons that generate three alternatively spliced mRNAs encoding three distinct protein isoforms.
Following the formation of a deoxynucleotide, the oxidized thiol group in RR have to be returned to its lowered state. The discount the RR thiol teams is carried out by both of two programs, the thioredoxin system and the glutaredoxin system. The last word supply of the electrons is NADPH. The thioredoxin system entails the protein recognized as thioredoxin (abbreviated Trx) and the enzymes recognized as thioredoxin reductases (abbreviated TrxR). The TrxR enzymes operate as homodimers and include a flavin (FAD) prosthetic group and possess a binding website for NADPH which is the terminal electron donor within the discount of RR.
Thioredoxin is encoded by the TXN gene. The TXN gene is positioned on chromosome 9q31.3 and consists of 5 exons that generate two alternatively spliced mRNAs encoding two distinct cytoplasmic protein isoforms.
People specific three thioredoxin reductase enzymes encoded by the TXNRD1, TXNRD2, and TXNRD3 genes. Every of the TXNRD encoded enzymes include selenocysteine residues which are included throughout their translation.
The TXNRD1 encoded protein is recognized as TrxR1. The TXNRD1 gene positioned on chromosome 12q23.3 and consists of 18 exons that generate seven alternatively spliced mRNAs encoding 5 totally different isoforms of TrxR1. The entire TXNRD1 encoded enzymes are cytoplasmic and are the principal enzymes concerned in deoxynucleotide synthesis.
The TXNRD2 encoded protein is recognized as TrxR2. The TXNRD2 gene positioned on chromosome 22q11.21 and consists of twenty-two exons that generate six alternatively spliced mRNAs leading to six totally different isoforms of TrxR2. Though all six TXNRD2 encoded enzymes are categorised as mitochondrial enzymes the isoform 3 and isoform 4 proteins lack the N-terminal mitochondrial focusing on sequence and are, due to this fact, predicted to be localized to the cytosol. The TXNRD2 encoded proteins, localized to the mitochondria, are primarily concerned in scavenging reactive oxygen species on this organelle.
The TXNRD3 encoded protein is recognized as TrxR3. The TXNRD3 gene positioned on chromosome 3q21.3 and consists of 16 exons that generate two alternatively spliced mRNAs leading to two totally different isoforms of TrxR3. The mRNA encoding the TrxR3 isoform 1 protein makes use of a non-AUG codon (CUG) to provoke translation. Just like the TXNRD1 encoded proteins, the TXNRD3 encoded proteins are concerned within the formation of deoxynucleotides.
The glutaredoxin system entails one in every of a number of proteins of the glutaredoxin household, the anti-oxidant peptide, glutathione (abbreviated GSH), and the enzyme glutathione reductase. Just like the TrxR enzymes, purposeful glutathione reductase comprises an FAD prosthetic group and an NADPH-binding website.
The glutaredoxins are a household of glutathione-dependent proteins that operate in a wide range of mobile redox reactions together with the formation of deoxynucleotides. People specific 5 genes that encode proteins containing the glutaredoxin purposeful area. 4 of the 5 proteins are referred to as glutaredoxins, the fifth protein is the enzyme recognized as prostaglandin E synthase 2 (encoded by the PTGES2 gene). The 4 glutaredoxin proteins are encoded by the GLRX, GLRX2, GLRX3, and GLRX5 genes.
The GLRX gene is positioned on chromosome 5q15 and consists of three exons that generate 4 alternatively spliced mRNAs, every of which encode the identical 106 amino acid cytoplasmic protein. The GLRX encoded protein is the first glutaredoxin concerned within the formation of deoxynucleotides.
The GLRX2 gene is positioned on 1q31.2 and consists of 5 exons that generate three alternatively spliced mRNAs that encode three distinct protein isoforms that possess distinct subcellular patterns of localization that embody the mitochondria, cytosol, and nucleus.
The GLRX3 gene is positioned on chromosome 10q26.3 and consists of 13 exons that generate three alternatively spliced mRNAs encoding two distinct protein isoforms. The first operate of the GLRX3 encoded proteins is within the regulation of the operate of a selected PKC isoform (PKCθ).
The GLRX5 gene is positioned on chromosome 14q32.13 and consists of two exons that encode a 157 amino acid precursor protein. The GLRX5 encoded protein is localized to the mitochondria the place it capabilities within the formation of iron-sulfur facilities in complexes of the oxidative phosphorylation pathway.
The glutathione reductase gene (image: GSR) is positioned on chromosome 8p12 and consists of 13 exons that generate 4 alternatively spliced mRNAs that encode 4 distinct mitochondrial protein isoforms.
Regulation of dNTP Formation
Ribonucleotide reductase is the one enzyme used within the technology of all of the deoxyribonucleotides. Due to this fact, its exercise and substrate specificity have to be tightly regulated to make sure balanced manufacturing of all 4 of the dNTPs required for DNA replication. Such regulation happens by binding of nucleoside triphosphate effectors to both the exercise websites or the specificity websites of the enzyme complicated. The exercise websites bind both ATP or dATP with low affinity, whereas the specificity websites bind ATP, dATP, dGTP, or dTTP with excessive affinity. The binding of ATP at exercise websites results in elevated enzyme exercise, whereas the binding of dATP inhibits the enzyme. Certainly, the binding of dATP dramatically decreases the exercise of RR in the direction of all 4 NDPs and explains, partly, the severely lowered deoxynucleotide manufacturing in ADA poor SCID. The binding of nucleotides at specificity websites successfully permits the enzyme to detect the relative abundance of the 4 dNTPs and to regulate its affinity for the much less ample dNTPs, to be able to obtain a stability of deoxynucleotide manufacturing.
Interconversion of the Nucleotides
Nucleoside Monophosphate Kinases
Through the catabolism of nucleic acids, nucleoside mono- and diphosphates are launched. The nucleosides don’t accumulate to any vital diploma, owing to the motion of nucleoside kinases. These embody each nucleoside monophosphate (NMPK) kinases and nucleoside diphosphate (NDPK) kinases.
The NMP kinases catalyze ATP-dependent reactions of the sort:
(d)NMP + ATP ↔ (d)NDP + ADP
There are 4 courses of NMP kinases that catalyze, respectively, the phosphorylation of:
- AMP and dAMP; household of kinases often known as adenylate kinases; 9 human adenylate kinase genes
- GMP and dGMP; household of two enzymes is aware of as guanylate kinases encoded by GUK1 and GUK2 genes
- CMP, UMP, and dCMP; household of two cytidine and uridine kinases (UCK1 and UCK2) that don’t phosphorylate the deoxynucleotides and a household of two kinases often known as cytidine/uridine monophosphate kinases 1 (CMPK1) and CMPK2 that phosphorylate the deoxynucleotides
- dTMP; household of two enzymes recognized as thymidine kinases (TK1 and TK2); the TK1 gene encodes a cytosolic enzyme, TK2 encode a mitochondrial enzyme that phosphorylates thymidine, deoxycytidine, and deoxyuridine essential for mitochondrial DNA synthesis
The adenylate kinase household of enzymes is vital for making certain ample ranges of vitality in cells corresponding to liver and muscle. The predominant response catalyzed by adenylate kinases is:
2ADP ↔ AMP + ATP
Nucleoside Diphosphate Kinases
The NDP kinases belong to a household of kinases recognized because the NME household. This nomenclature stems from the identification of the NM23 (non-metastatic clone 23, also referred to as NME) gene whose encoded protein was initially recognized as being liable for metastasis suppression within the murine melanoma mannequin system. The NM23 gene was discovered to encode one of many subunits of NDPK, recognized as NDPK A. The human NME/NM23/NDPK household consists of 9 members recognized as NME1-NME9.
The varied members of the NDPK household catalyze reactions of the sort:
N1TP + N2DP ↔ N1DP + N2TP
N1 can characterize a purine ribo– or deoxyribonucleotide; N2 a pyrimidine ribo– or deoxyribonucleotide. The exercise of the NDP kinases can vary from 10 to 100 occasions greater than that of the NMP kinases. This distinction in exercise maintains a comparatively excessive intracellular degree of (d)NTPs relative to that of (d)NDPs. In contrast to the substrate specificity seen for the NMP kinases, the NDP kinases acknowledge a large spectrum of (d)NDPs and (d)NTPs.
Along with the position of NDPK in sustaining the pool of NTP and dNTP for nucleic acid (DNA and RNA) homeostasis, these enzymes present a pool of CTP utilized within the technique of phospholipid synthesis, UTP utilized within the synthesis of glycoconjugates corresponding to glycoproteins, and GTP required for GPCR-mediated sign transduction.
