Growth of MVsim
MVsim is a multivalent interplay toolset constructed upon our configurational microstate community mannequin23, which expanded upon prior modeling efforts within the literature by explicitly treating multivalency as a dynamic ensemble of binding configurations pushed by means of native, topology-derived efficient concentrations. MVsim represents a reconceptualization and utility of the preliminary, extra restricted community mannequin to now present mechanistic descriptions of an array of biologically and therapeutically related multivalent methods and to quantitatively predict binding responses and conformational dynamics throughout a breadth of parameter area31,37,40.
The creation of the MVsim toolset translated the elemental ideas of the community mannequin into the MATLAB coding setting by means of a collection of implementations that signify important advances in its capacity to simply simulate a broad vary of multivalent interactions. First, to explain a user-specified multi-ligand, multivalent interplay system, we developed a rule-based modeling routine to routinely enumerate all attainable binding microstates and configuration transitions between them in an effort to generate a descriptive kinetic mannequin (Prolonged Strategies, Supplementary Info). For instance, prolonged to its furthest, MVsim simulates aggressive interactions amongst three topologically-varied trivalent ligands for a trivalent receptor, described with a system of 1538 differential fee equations.
Second, MVsim successfully and quickly parameterizes the system of fee equations with computed topology-derived first-order fee constants of affiliation. Right here, MVsim makes use of dimensionally-reduced polar coordinate integrations of the molecular interplay volumes. With this method, the frequency of all pairwise mixtures of multivalent interplay between a ligand and receptor binding area are calculated with joint likelihood density capabilities to yield a set of efficient concentrations. This routine permits environment friendly calculations to be carried out with excessive spatial decision for nanoscale and mesoscale multivalent species with area diameters, linkages, and persistence lengths exceeding 1000 Å.
Third, MVsim has an intensive multiparameter description of the molecular multivalent panorama that permits for zero-fit prediction of the response dynamics of absolutely parameterized methods the place experimental multivalent information are absent. Conversely, MVsim permits parameter estimation for topologically under-characterized methods the place multivalent binding kinetics have been measured. MVsim facilitates quantification of multivalent binding responses by way of efficient fee constants of affiliation (({ok}_{{{rm{on}}}}^{{{rm{eff}}}})) and dissociation (({ok}_{{{rm{off}}}}^{{{rm{eff}}}})), equilibrium dissociation constants (({Ok}_{{{rm{D}}}}^{{{rm{eff}}}})), aggressive inhibitor efficiency (({{IC}}_{50})), and Hill coefficients (nH) describing ultrasensitive switch-like habits.
Parameter inputs
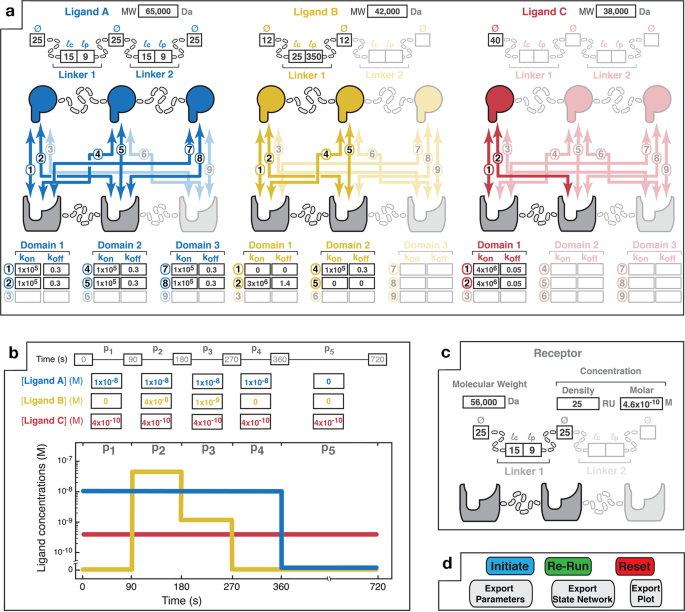
Interfacing MVsim with MATLAB’s app design setting enabled the creation of a tabbed GUI to information the specification of biologically necessary manifestations of multivalency by way of multiparameter inputs. The MVsim GUI permits full enter parameterization of the area and linkage topologies of the ligand(s) (Fig. 1a) and receptor (Fig. 1c), the monovalent kinetics between every pairwise mixture of ligand-receptor binding domains (Fig. 1a), the temporal ligand focus dynamics (Fig. 1b), and the set of parameters that govern SPR and associated kinetic research, together with the affiliation and dissociation occasions, circulate fee, and stage of the immobilized receptor. As well as, for cases the place detailed topological info is understood, MVsim permits customers to immediately enter efficient ligand concentrations and end-to-end likelihood density capabilities for the multivalent system of curiosity (Supplementary Fig. 1). Lastly, as soon as a multivalent design has been inputted, the person can provoke MVsim and subsequently survey a spread of non-topological parameter variants in fast succession (Fig. 1d). Tutorials for navigating the GUI and inputting parameters can be found within the Supplementary Info and on the MVsim GitHub web page (https://sarkarlab.github.io/MVsim/). Supporting figures that element the parameter inputs used for every simulation on this research are additionally supplied within the Supplementary Info.
a Some extent-and-click interface permits the person to pick out the variety of ligands (as much as three) and valencies of the ligand(s) and receptor (as much as trivalent) that compose the multivalent system. Primarily based upon the chosen design, the person specifies the construction of every of the ligands by getting into the relevant molecular weight (MW); the binding area diameters (Ø); the contour lengths (({l}_{{{{{{rm{c}}}}}}})) of the linkers (i.e., the utmost end-to-end distance; e.g., 3.5 Å and 1.5 Å per amino acid for a random coil and alpha helix, respectively); and the persistence lengths (({l}_{{{{{{rm{p}}}}}}})) of the linkers. Additional, the relevant combinatorial interactions (numbered 1 to 9) distinctive to every receptor–ligand pairing are highlighted. Parameter fields enable the enter of monovalent fee constants for every pairwise interplay. Non-binding interactions will be indicated with okon and okoff values of zero (e.g., as illustrated with Ligand B in yellow for interactions “1” and “5”). b An enter area permits the person to specify patterns of the whole, bulk ligand concentrations. An affiliation part happens during times of non-zero bulk ligand focus (e.g., 90–270 s for Ligand B). Dissociation phases happen when the ligand is faraway from the majority resolution (e.g., 360–720 s for Ligand A). Right here, Ligand C is specified as repeatedly current in resolution through the 720 s of the interplay timecourse. The graphical show permits visualization of the desired bulk focus pulse sample. c Consumer enter parameters for the receptor. Receptor focus will be specified as both an SPR-mimicking floor density (measured in RU; the place 1 RU equals ~1 pg/mm2) or a molar focus. Receptor topology is laid out in the identical kind as described above for the ligands. d The MVsim controller tab permits initiation, iteration, and export of binding simulations. “Provoke” executes a simulation. “Re-run” executes an abbreviated simulation used when no adjustments had been made to the valency or topology of the system. “Reset” relaunches the app and clears person enter parameters from all fields. Tutorials illustrating the enter interfaces can be found within the Supplementary Info and at https://sarkarlab.github.io/MVsim.
Simulation outputs
Following the initiation of a simulation, MVsim gives customers quite a lot of means to visualise, work together with, and export the simulated response kinetics. Most easily, the simulation outcomes are displayed throughout the output area as an interactive plot of the binding response sign as a perform of the desired affiliation and dissociation time (Fig. 2). Right here, customers can select between two graphical outputs of the response kinetics. First, as is typical of experimental kinetic binding information, a plot of a user-specified ligand focus is displayed (Fig. 2a). Alternatively, customers can choose a plot of all composite microstate configurations underlying the response sign (Fig. 2b), binned in response to valency class (Fig. 2c) or by ligand class to observe the aggressive binding dynamics amongst a number of ligands (Fig. 2nd). MVsim moreover permits customers to visualise the dynamic evolution of the microstate community by way of an interactive map (Fig. 2e) and to export the response kinetics as a set of tab-delineated textual content recordsdata to facilitate deeper evaluation by means of offline plotting and curve becoming, and thru microstate community evaluation throughout the Cytoscape software program setting41. MVsim additionally permits customers to immediately examine each the computed likelihood density capabilities and efficient concentrations (Supplementary Fig. 2).
a A simulated SPR sensorgram shows the general response dynamics (i.e., summation of all ligands and binding microstates) for specified ligand focus(s). Indicated listed here are the binding responses for a serial dilution of a single ligand binding to a receptor with affiliation (0–300 s) and dissociation (300–600 s) phases. For a easy quantitative comparability between simulations, an general efficient OkD will be calculated by the equilibrium methodology. b For a specified ligand focus, all composite microstates are displayed. c, d To facilitate analyses of the binding responses, the microstates will be binned in response to both c valency or d ligand class. e For visible evaluation of the evolution of a community of microstates in b, an interactive graph exhibits inhabitants adjustments in microstate lessons over a timecourse of affiliation and dissociation. The simulations in these determine panels aren’t associated to 1 one other; they’re for illustrative functions to spotlight the distinctive options in every view. Tutorials illustrating the output interfaces and options can be found within the Supplementary Info and at https://sarkarlab.github.io/MVsim.
Assessing efficiency in opposition to experimental mannequin methods
To judge the accuracy of simulating advanced topologies, MVsim was used to foretell the binding response dynamics for 3 multivalent methods (Fig. 3). First, we evaluated our beforehand constructed monospecific multivalent interplay (i.e., composed of a single pair of protein interplay domains; Fig. 3b). Then, we evaluated two further experimental methods: a multispecific multivalent interplay (i.e., composed of two units of interplay domains; Fig. 3d) and a mixed multispecific/multi-ligand interplay (Fig. 3g). To immediately examine MVsim to our earlier research, we evaluated a monospecific interplay composed of a kinetically and structurally parameterized pair of protein–protein interplay domains (SH3 and SH3-binding peptide (SBP); our experimental parameterization by SPR is proven in Fig. 3a)42, rendered multivalent with polypeptide linkages parameterized utilizing literature-derived topological values (Fig. 3b)43,44,45,46. Right here, MVsim predicts monospecific multivalent binding (Fig. 3c) however now additionally exhibits improved sensitivity to the topological constraints that may impede sure configurations, corresponding to people who requiring contorted twisting of interdomain linkages, leading to model-experiment settlement with a root-mean-square error (RMSE) of two.1 RU (7% of the common sign; Fig. 3c). Furthermore, by extending these simulations by means of systematic parameter variation, MVsim recognized ligand focus because the parameter with probably the most delicate impact on the simulated binding response of this method. By growing the simulated ligand focus simply twofold, an improved general model-experiment settlement was noticed in each the low focus ligand circumstances and within the magnitude of the biphasic affiliation. This enchancment in match, nevertheless, is on the expense of capturing the modest kinetic burst at early occasions (Supplementary Fig. 3a), underscoring the advanced, interconnected relationships between a single parameter worth and a number of descriptive noncanonical options of multivalent binding responses.
a Monovalent SPR kinetic fee constants had been experimentally decided for the SH3-binding peptide (SBP)-SH3 (a, left panel) and Prb-Pdar (a, proper panel) interactions that had been used to construct the multivalent methods. Kinetic suits with a “fast mixing” 1:1 Langmuir mannequin confirmed good settlement for the reason that experimental circumstances weren’t considerably mass-transfer restricted (see Supplementary Experimental Strategies). b A trivalent, monospecific receptor–ligand interplay was engineered and parameterized inside MVsim utilizing values for the kinetic fee constants of affiliation (okon) and dissociation (okoff), area diameters (Ø), and contour (lc) and persistence (lp) lengths for the linkers. c Simulated (c, left panel) and experimental (c, proper panel) binding response dynamics for the trivalent, monospecific interplay at seven ligand concentrations (0.98, 3.9, 15.6, 62.5, 250, 1000, and 2000 nM). Mannequin-experiment RMSE is 2.1 RU. d A trivalent, bispecific receptor–ligand interplay was engineered and parameterized utilizing values for the kinetic fee constants of affiliation (okon) and dissociation (okoff), diameters (Ø) for the protein–protein binding domains, and contour (lc) and persistence (lp) lengths for the alpha-helical linkers. Arrows point out suitable interactions between receptor and ligand binding domains. e Simulated binding responses for the parameterized trivalent, bispecific interplay at 4 simulated ligand concentrations (0.1, 1, 10, and 1000 nM). f Experimental SPR binding response dynamics for the trivalent, bispecific interplay on the identical 4 ligand concentrations as in e. Mannequin-experiment RMSE is 1.6 RU. g The Pdar-Prb and SBP-SH3 protein–protein binding domains had been used to create a multi-ligand system. h Simulated binding response dynamics modeled by MVsim for the parameterized twin ligand system. An overlay is proven of binding responses for 3 simulated mixtures of ligands A and B (1 nM A + 2.5 nM B; 1 nM A + 50 nM B; and 1 nM A + 250 nM B). i Experimental SPR binding response dynamics for a similar three twin ligand mixtures as in h. Mannequin-experiment RMSE is 2.0 RU. The enter parameters for these simulations are supplied in Supplementary Fig. 5. Supply information are supplied as a Supply Knowledge file.
We additional used MVsim to make predictions of multispecific and multi-ligand interplay methods. Within the first validation, a multispecific receptor–ligand structure was designed by means of incorporation of a second set of protein–protein interplay domains (Prb and Pdar; Fig. 3a). Once more, along with the experimentally decided monovalent kinetic fee constants (Fig. 3a), literature-derived properties of the molecular buildings and topologies had been used to parameterize the mannequin (Fig. 3d)43,44,45,46 and to generate a simulated dataset (Fig. 3e). Evaluating simulation with the corresponding SPR dataset (Fig. 3f) demonstrates good settlement with regard to the power of MVsim to foretell a priori the magnitude and multiphasic character of the experimental binding responses. Additional, MVsim gives mechanistic explanations for these binding responses, exhibiting, for instance, the contribution of high-stoichiometric configurations to the microstate ensemble pushed by way of inflexible, α-helical linkers (Supplementary Fig. 4a–c). The quantitative RMSE between mannequin and experiment of 1.6 RU (6% of the common sign) signifies that the simulations successfully captured the experimental binding responses to a big diploma. Transferring past zero-fit predictions, MVsim identifies parameters that almost all sensitively have an effect on the multivalent binding response. Right here, for instance, the persistence lengths of the ligand linkages current as probably the most delicate system parameters. Will increase to the rigidity of the ligand linkers on this system served to put the ligand-receptor binding domains barely out of optimum register, modestly enhancing the presence of high-stoichiometric binding states within the affiliation part and growing the speed of dissociation (Supplementary Fig. 3b).
As a second model-experiment validation, the monospecific and multispecific designs (Fig. 3a, b, d) had been mixed to create a kinetically and topologically parameterized twin ligand interplay system (Fig. 3g). The simulated binding responses (Fig. 3h) reach capturing the multiphasic affiliation and dissociation dynamics current within the experimental SPR information (Fig. 3i). Furthermore, past merely predicting the general kinetics of the system, MVsim gives insights into the mechanics of a number of multivalent and multispecific ligands competing for a receptor, and attributes these molecular properties again to the macroscopically observable options of the multiphasic binding responses. Right here, for instance, MVsim captures how efficient fee constants of dissociation will be dictated by valency and can be utilized to impact the temporal ordering of interactions between a quickly, however extra transiently, binding monovalent ligand and a slower, however extra avid, multivalent ligand (Supplementary Fig. 4d–f). Additional, as within the multispecific validation, parameter variation can once more be used to evaluate model-experiment settlement. Right here, for instance, our zero-fit simulation of this multivalent, bispecific, and multi-ligand interplay system agreed with the experiment with an RMSE of two.0 RU (9% of the common binding sign). Whereas the zero-fit simulation once more captured the biphasic options of the affiliation and dissociation phases of the binding response, the relative proportions of the quick and gradual phases had been much less effectively predicted. Via parameter variation, MVsim recognized ({ok}_{{{rm{on}}}}) for Ligand B as a parameter that sensitively impacts these multiphasic binding responses (Supplementary Fig. 3c). Right here, for instance, a twofold improve within the Ligand B ({ok}_{{{rm{on}}}}) within the mannequin yielded higher quantitative settlement to the quickest part of the experimental affiliation curve on the highest ligand focus (Supplementary Fig. 3c; 0–2 s of the affiliation part) and the slowest part of the experimental dissociation curve (Supplementary Fig. 3c; 125–250 s). Nonetheless, these two enhancements in model-experiment settlement got here on the expense of over-representing the quantity of Ligand B that continues to be certain to the receptor at equilibrium (Supplementary Fig. 3c; ~125 s). Once more, this evaluation of parameter sensitivity highlights the advanced relationship between a single parameter worth and the descriptive options of the binding response, right here particularly in regard to the aggressive dynamics between the ligands for the receptor.
Functions to multivalent system design and quantification
MVsim was established to each information the design and implementation of multivalent properties and to facilitate parameter estimation for present and incompletely characterised pure and artificial multivalent methods. Right here, the mannequin’s lack of reliance upon fitted parameters permits MVsim to higher describe the additive, aggressive, and cooperative relationships implicit amongst kinetic, topological, and valency parameters and to use these to the quantification of multivalent properties, corresponding to efficient focus, avidity, and binding selectivity. To judge the efficiency of MVsim as a molecular design and quantification software, we assessed its capacity to design and predict binding response dynamics in 4 totally different cases and functions of multivalency.
MVsim predicts ultrasensitive habits in engineered protein switches and logic gates
The efficient focus that drives multivalent binding offers these methods the inherent capacity to supply nonlinear enter/output response dynamics. It has been beforehand demonstrated, for instance, that ultrasensitive toggling will be pushed by means of the introduction of monovalent counterparts right into a multivalent system31. Dueber et al. confirmed that cooperative aggressive dissociation of multivalent protein–protein complexes results switch-like transitions that may be leveraged to regulate the fractional saturation of receptor–ligand interactions and enzymatic exercise. Right here, we apply MVsim to review the activation dynamics of engineered bivalent and trivalent protein switches and establish important parameters for optimum system efficiency. MVsim quantitatively predicts the connection between the valency of the system and the magnitude of its cooperative transition to an energetic state (Fig. 4a). The practical vary of multivalent switches will be prolonged by means of the incorporation of multispecific interactions. This design method permits the creation of AND logic gates by which a swap response is elicited solely by a programmed mixture of molecular inputs. Though catalytic exercise (and never SPR) was used to measure output on this experimental system, MVsim may nonetheless qualitatively seize the three-input gating perform for the obtainable experimental information (Fig. 4b). As well as, our simulations recommended doubtless spurious two-input activation of the system (Supplementary Fig. 6a, b; bars v–vii), as a consequence of the nontrivial activation that’s noticed computationally by single inputs (Supplementary Fig. 6b; pink bars ii–iii) and that’s much more outstanding within the experiments, as a result of important basal activation and single-input PDZ activation (Supplementary Fig. 6b; grey bars i and iv, respectively). The simulations did not predict these latter two cases, which had been the mildest activators (i.e., no enter or the lowest-affinity enter), suggesting that the sterically constrained experimental system produces basal activation past the idealized topological remedy within the simulations. Lastly, by means of parameter exploration, MVsim guided the identification of key steric constraints throughout the system that would contribute to the noticed basal and single-input activations and indicated an optimized molecular design (Supplementary Fig. 6c) that would decrease these impediments throughout the auto-inhibited conformation with prolonged versatile linkages and equalized the interdomain binding kinetics to enhance the dose-responsiveness of the agonists. The outcome was a extra tightly managed simulated system that higher maintains an inhibited state within the presence of zero-, one-, and two-input circumstances (Supplementary Fig. 6c; bars i, ii–iv, and v–vii, respectively).
a Experimental response dynamics of artificial monovalent and trivalent switches from ref. 31 had been used to benchmark the predictive efficiency of MVsim simulations described by the reported structural, topological, and kinetic parameters. Ultrasensitivity of every simulated response is reported with a calculated Hill coefficient (nH) for direct comparability with the reported literature values. b Experimental output responses for a trispecific AND logic gate, additionally from ref. 31, benchmarked in opposition to an identically parameterized system in MVsim. For readability, the AND gate is depicted (diagram, prime left) in its initially designed twisted configuration (detailed in Supplementary Fig. 6). Simultaneous addition of the three inputs (SH3, PDZ, and GBD-binding peptides; coloured purple, yellow, and blue, respectively) flips the AND gate into an energetic conformation. To make sure optimum system efficiency, it’s fascinating to stop activation of the AND gate with subthreshold inputs, although that is troublesome to attain by means of advert hoc experimentation. Right here, nontrivial subthreshold activation is certainly noticed (bars i–iv). c MVsim specifies optimum design of multivalent and multispecific ligands to yield desired patterns of selective interactions inside a pool of three receptors with frequent binding domains. The affinities for the receptor–ligand binding domains (coloured blue, yellow, and purple) are as described in b. The MVsim enter parameters for the simulations in a–c are additional detailed in Supplementary Fig. 6.
MVsim informs using multispecificity for molecular recognition and therapeutic focusing on
Multispecificity is a potent molecular design ingredient that’s extensively utilized in drug discovery and cell engineering. By leveraging two or extra distinct binding epitopes, multispecific interactions are employed to engineer extremely avid and selective molecular recognition to be used in such functions as bispecific therapeutic antibodies10,27 and chimeric antigen receptor T cells47. Multisite recognition moreover permits higher-order info processing, permitting these multispecific methods to generate differential outputs to various mixtures of inputs11,12. As a result of the community mannequin of multivalency computes multivalent binding because the cooperative sum of its composite interactions, MVsim is well-suited to the research of such multipartite interactions.
For instance, multispecific interactions will be designed to maximally exploit any diploma of variation within the kind and variety of floor receptors and antigens inside a inhabitants for the needs of selective focusing on10. On this regard, we directed MVsim to handle a design query: given a inhabitants of three distinct kinds of antigenic cell surfaces (Fig. 4c), what are the optimum ligand designs that may singly, doubly, and triply interrogate the inhabitants? MVsim demonstrates that the composition of the goal receptor serves as a typically helpful information for ligand design, as seen, for instance, within the relative selectivites of mono-, bi-, and trispecific Ligands 1, 3, and seven, respectively, for Receptor 3 (Fig. 4c). Furthermore, selective recognition will be additional tuned utilizing designed linkages that leverage the spatial proximity between receptor goal surfaces; Ligand 2b (inflexible linkage) has higher selectivity than Ligand 2a (versatile linkage) for Receptor 2 (Fig. 4c).
The knowledge-coding capability of multivalent interactions may also impact the temporal ordering of ligand binding to a single receptor goal when a number of multivalent ligands are launched concurrently (Supplementary Fig. 7a–c), a phenomenon that isn’t attainable in a comparable monovalent system (Supplementary Fig. 7d–f). Exploration of the simulated parameter area on this system revealed a multivalent design leveraging kinetics, avidity, and stoichiometry that allows serial phases of dominant ligand engagement by exploiting the cumulative results of concurrent binding afforded by multispecificity, the cooperative, aggressive binding of multi-ligand dynamics, and the technology of efficient dissociation fee constants by way of multivalency (Supplementary Fig. 7c).
MVsim fashions the multivalency and avidity of SARS-CoV-2 S protein interactions
At current, some of the outstanding and consequential shows of multivalent binding entails the floor spike (S protein) of SARS-CoV-2. The S protein is a classy, conformationally activated molecular system that mediates selective recognition of goal cells and generates the driving drive wanted to beat the vitality barrier of membrane fusion, thus enabling viral entry into the host32,33. The multimeric and multivalent configuration of the S protein is central to those capabilities48. Trimeric meeting serves to stabilize the S protein in opposition to faulty fusogenic conformational adjustments, set up allosteric management, and doubtlessly current a number of receptor binding domains (RBDs) that bind multivalently with a host-cell floor populated with dimeric ACE2 receptor proteins48. In response to those pure shows of multivalency, this identical precept has been mobilized in therapeutic designs supposed to neutralize, inhibit, or in any other case uncouple the construction–exercise relationship of the S protein35,36,37,38,39,40.
Regardless of the considerably extra advanced multivalent structure of the S protein in contrast with our beforehand described functions, MVsim will be successfully parameterized to mannequin and quantify important structural properties of the S protein-ACE2 interplay (Supplementary Fig. 8a, b). For instance, it stays an open query the extent to which the trimeric S protein can multivalently interact a bivalent ACE2 receptor. That is of appreciable significance for our understanding of how the affinity and avidity of S protein binding relate to infectivity, and what penalties this poses for therapeutic inhibition40,49. In synthetically engineered multivalent cases of the RBD-ACE2 interplay, MVsim quantitatively predicts the relative lack of steric hindrance that affords the ultra-high-avidity binding noticed in a research by Chan et al.40 (Fig. 5a, b). In distinction, experiments carried out on extra biologically mimetic S protein-ACE2 interactions point out a big obstacle towards high-avidity binding40. Utilizing MVsim to suit therapeutic neutralization datasets reveals an efficient ligand focus, ([{L}_{{{rm{eff}}}}]), for the second engagement occasion between S protein and ACE2 that’s 2000-fold much less potent than that noticed within the sterically unimpeded system (Fig. 5c, d)40. This incapability to attain high-avidity binding (e.g., a community by which >95% of the populated microstates are certain with maximal valency, as is the case for the “Excessive” simulation in Fig. 5d) will be defined by the mixture of the rigidity of the ACE2 dimer and the apparently constrained, directional movement of the linkage tethering the RBD. Quantitative modeling approaches corresponding to these point out a big potential for therapeutic designs that may potently outcompete the RBD-ACE2 interplay by leveraging multivalent binding in methods inaccessible to the S protein (Fig. 5e, f). Particularly, MVsim predicts that as much as 1000-fold enhancements in IC50 values will be achieved by means of using topologically exact and constrained linkages inside a designed, trivalent multispecific neutralizing therapeutic (Fig. 5e, f). Reciprocally, MVsim additional demonstrates how bivalency will be successfully leveraged with applicable linkages to avidly block the RBD binding surfaces of the ACE2 dimer (Supplementary Fig. 8c, d).
a An idealized, versatile artificial design of an ACE2–RBD bivalent structure. Right here, the artificial design removes the RBD from the biologically related and constrained context of the remainder of the S protein. b The versatile RBD linkers afford a high-avidity bivalent interplay with the dimeric ACE2 that was past the quantification limits of the experimental SPR. Right here, MVsim was parameterized with the options of the experimental methods and gives prediction and quantification of the ultra-high-avidity interplay. c Utility of MVsim to a biologically related occasion of the SARS-CoV-2 S protein RBD and ACE2 interplay. Right here, the therapeutic neutralizing exercise of soluble, dimeric ACE2 (purple) was quantified in a SARS-CoV-2 pseudovirus-host-cell system40. d The ensuing IC50 datasets had been utilized to MVsim in an effort to match for a extra biologically related willpower of the multivalent binding capability of the S protein-ACE2 interplay. The experimental information (purple traces) are tailored from ref. 40 The perfect match from MVsim gave an [Leff] of 100 nM (curve outlined in black), falling between the “Low” and “Medium” commonplace curves (shades of pink), which signify no capability and a modest steric capability for bivalent binding, respectively. These simulations point out that the RBDs within the full context of the S protein are considerably impeded for direct bivalent binding to ACE2. e This steric obstacle will be exploited to maximise neutralizing efficiency by absolutely leveraging therapeutic multivalency. f MVsim can take a look at the design of neutralizing inhibitors that maximally outcompete the ACE2 interplay. Designs leveraging monospecific bivalency (prime panel) and trivalent bispecificity (backside panel) are computationally modeled for his or her neutralizing strengths and off-rate dependent pharmacokinetic half-lives within the presence of fixed S protein (orange bar above plot) and decaying focus of therapeutic (blue bar). The enter parameters for these simulations are given within the MVsim person tutorial within the Supplementary Info.
MVsim quantifies the dynamics of SARS-CoV-2 S protein conformational switching
Along with the sterically impeding immobility of the S protein-ACE2 interplay (Fig. 5c, d), multivalent engagement is proscribed by the accessibility of the RBDs, as they dynamically pattern configurations starting from the occluded but stabilized “RBD-down” conformation to the labile but ACE2-binding competent “RBD-up” conformation33,48. The dynamics of this vary of RBD movement are a big goal of selective strain as the advantages of maximizing host-cell binding are countered by the necessity to stabilize the S protein in opposition to spontaneous fusogenic conformational change and immune surveillance of uncovered important surfaces48. To look at intramolecular conformational adjustments that yield multivalent binding, we utilized MVsim to simulate a multicomponent experimental system consisting of a stabilized trivalent S-protein, a set of first-order fee constants (({ok}_{{{rm{up}}}}) and ({ok}_{{{rm{down}}}})) describing RBD conformational change, and a trivalent ligand particular for the RBD-down conformation (Fig. 6a). MVsim, constructed and parameterized on this means, succeeded not solely in recapitulating the experimental multiphasic kinetic traces obtained in a research by Schoof et al.37 (Fig. 6b, c), but additionally in relating these response dynamics to the charges of RBD conformational switching. MVsim was used to derive the best-fit parameter values that describe the conformational switching: ({ok}_{{{rm{up}}}}) = 0.017 s−1 and ({ok}_{{{rm{down}}}}) = 0.008 s−1 (Fig. 6d, e). These correspond to particular person RBD half-lives of ~1.4 min within the RBD-up configuration and ~0.7 min within the RBD-down configuration for this stabilized, in vitro S protein system. The info-richness of multiphasic SPR sensorgrams is underscored by the truth that the S protein binding response dynamics are uniquely decided by a single set of kinetic constants (Fig. 6d, e). For instance, variations in ({ok}_{{{rm{up}}}}) and ({ok}_{{{rm{down}}}}) that keep the equilibrium fixed (i.e., fixed ({ok}_{{{rm{up}}}}/{ok}_{{{rm{down}}}}) ratio) nonetheless end in diagnostically totally different response dynamics (Supplementary Fig. 9). To additional assess the accuracy of the MVsim-derived values of ({ok}_{{{rm{up}}}}) and ({ok}_{{{rm{down}}}}), these fee constants had been used to parameterize a simulated SPR experiment probing the lifetime of the stabilized RBD-all-up state. Right here, good settlement was noticed between mannequin and experiment (Fig. 6f, g)37.
a MVsim fashions the conformational change as an intramolecular ligand binding occasion (coloured in grey) that toggles the trivalent S protein between “RBD-up” and “RBD-down” conformations. The conformational change is outlined by a pair of first-order kinetic fee constants okdown and okup. b, c The experimental kinetics of conformation-specific nanobody binding, tailored from ref. 37 (b), are qualitatively predicted by a zero-fit simulation with MVsim (c). d, e Making use of a gradient descent becoming routine to a few of the six experimental sensorgrams in b ([nanobody] = 3.13, 12.5, and 50 nM; the second, fourth, and sixth curves from the underside, respectively) that minimizes the root-mean-square error (RMSE) between mannequin and simulation throughout a broad vary of values for okdown and okup (d, prime panel) converges on a novel resolution for a single set of best-fit parameter values for okdown (d, backside left panel) and okup (d, backside proper panel) for a person RBD (e). f Conformational switching half-life experiments additionally tailored from ref. 37, alter the relative proportion of “gradual part” inhibitor dissociation occasions (i.e., high-avidity bivalent and trivalent interactions) and “quick part” inhibitor dissociation occasions (i.e., monovalent interactions). Right here, as a result of comparatively gradual RBD switching charges, longer affiliation occasions allow extra S protein to be certain in high-avidity interactions, and thus give rise to small percentages of “quick part” dissociation occasions. g To evaluate MVsim accuracy, the fitted parameters are utilized in a modeling framework to simulate the experimental system and examine half-lives (t1/2) of “RBD-up” S protein conformations. The enter parameters for these simulations are given within the MVsim person tutorial within the Supplementary Info.
Given the distinctiveness of the fitted parameters on this system, MVsim ought to be equally able to uncovering the conformational dynamics of different S proteins. Notably, the half-lives for the synthetically stabilized S protein examined on this research are ~35-fold slower than these just lately measured for the native S protein utilizing FRET sensors50. As well as, S protein conformational dynamics are of specific significance for understanding the mechanisms by means of which rising SARS-CoV-2 mutational variants of concern (VOCs) improve infectiousness51. Mechanistically, S protein VOCs can perform to stealth this protein from host immune surveillance, improve the binding kinetics/affinity of the RBD-ACE2 interplay, stabilize the RBD-up configuration to extend the avidity of virion-host-cell engagement, and/or increase conformational allostery that allows RBD binding to prime activation of membrane fusion51. To additional apply MVsim to review S protein conformational dynamics, simulations had been carried out with the parameterized S protein RBD ensemble (Supplementary Fig. 10a) to probe the results of ({ok}_{{{rm{up}}}}) and ({ok}_{{{rm{down}}}}) on the ensemble of RBD configurations (Supplementary Fig. 10b–d). Stabilization of the RBD-up state results in commensurately stronger ACE2 receptor binding (Supplementary Fig. 10e–g).