Pressure ON4T genome sequencing
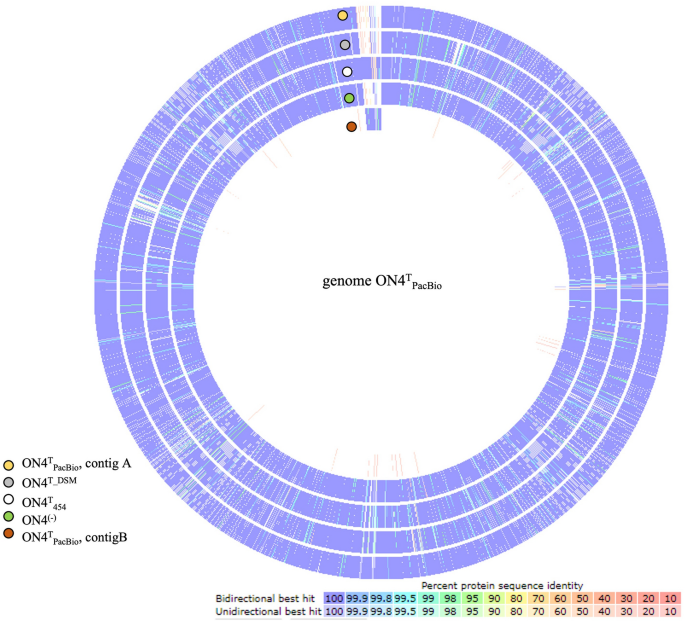
The 454 pyrosequencing of the G. molinativorax ON4T genome (ON4T454) generated 305,068 reads, comprising 133.7 million nucleotides (nt), which had been assembled in 163 contigs (PXVE00000000). Considering disclosing the genes within the neighborhood of molA, analyses began with a BLAST search of the molA gene sequence in opposition to the 163 assembled contigs. The whole molA gene sequence (1398 bp) was positioned in a single contig (1957 bp) however revealed no different open studying body (ORF) in its neighborhood. The pressure ON4T genome was resequenced utilizing PacBio (ON4TPacBio), which generated 102,855 polymerase reads, leading to 184,306 subreads with a imply subread size of 7254 bp, and 1,337 billion nt. These reads had been de novo assembled and two contigs of three,465,658 bp (linear) (contig A—CP028426) and 37,013 bp (round) (contig B—CP028427) had been generated. The assembled genome had 98.6% completeness, 0.6% contamination (Desk S1) and 64% common G+C content material for every contig. This in silico G+C content material worth agrees with beforehand revealed knowledge7. The ON4TPacBio genome meeting disclosed 3243 predicted protein-coding genes, most of them assigned to putative features (75.9%) and matching 94.3% of the anticipated protein-coding genes within the meeting of pressure DSM13485T (JGI, AUDX00000000.1) (Desk 1, Fig. 1, Desk S1). Moreover, forty-nine tRNAs, three 16S rRNA, three 23S rRNA, and three 5S rRNA genes had been annotated (Desk 1). The prevalence of three 16S rRNA gene copies was additional confirmed by figuring out the ratio between the 16S rRNA and the recA genes (single-copy gene) in each strains ON4T and ON4(−) (Desk S2).
As anticipated, the information retrieved utilizing each sequencing applied sciences (454 and PacBio) had been very comparable (Fig. 1). Nonetheless, the lengthy reads obtained by PacBio sequencing allowed to sum up the 163 assembled ON4T454 contigs into two contigs solely. As well as, the comparability of the 2 draft genomes allowed closing the contig A (Fig. S1) and figuring out the molA gene within the contig B.
molA is the passenger gene of Tn6311
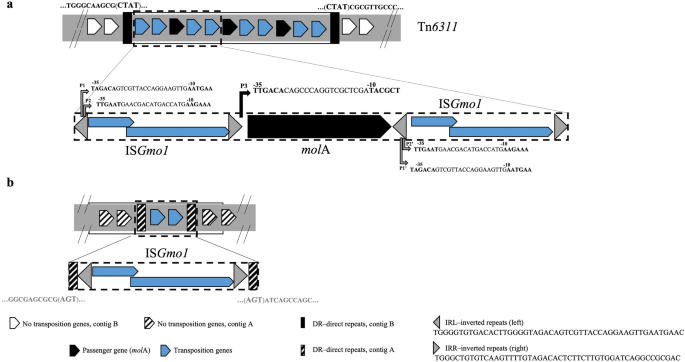
The molA gene, positioned in contig B, was surrounded by two equivalent insertion sequences (IS) of the IS3 group and household (Fig. 2a), whose unique transposase belongs to the DDE transposase household26. The first sequence of this IS was not equivalent to any IS described to this point and was named ISGmo1 (https://www-is.biotoul.fr/). This construction, the place a passenger gene (molA), which doesn’t code for a transposition enzyme, is flanked by two IS (ISGmo1), is typical of a composite transposon27,28. This group was confirmed by Sanger sequencing (knowledge not proven). The novel composite transposon, named Tn6311, contained three copies of molA, every flanked upstream and downstream by ISGmo1 (a complete of 4 ISGmo1 copies) (Fig. 2a). This tandem group was primarily noticed in particular PacBio uncooked reads (knowledge not proven). Additional, the three molA copies had been confirmed in pressure ON4T utilizing quantitative PCR, as indicated by the gene copy ratio between molA and a neighbour single-copy gene (parA) (Desk S2).
Mapping of the molA gene genetic neighborhood. (a) Illustration of the composite transposon Tn6311 in pressure ON4T (ON4TPacBio—CP028427), in addition to the in silico predicted promoters (P) near molA gene. (b) Schematic illustration of the ISGmo1 with out passenger gene in pressure ON4T and ON4(−) (ON4TPacBio—CP028426; and ON4T454—PXVE00000000; DSM13485T (= ON4T)—AUDX00000000.1 and ON4(−)—PXVD00000000, http://www.ncbi.nlm.nih.gov/).
As described for different IS3-like sequences, ISGmo1 has a complete size of 1287 bp and comprises two partially overlapping open studying frames, orfA (291 bp) and orfB (1080 bp), which encode the DNA-binding and the catalytic area of the transposase, respectively27,29,30,31. Some IS3 relations may assemble completely different useful protein domains from one DNA section, utilizing a mechanism referred to as recoding programmed ribosomal frameshifting. This mechanism entails a ribosomal slippage of 1 nt, occurring on the heptanucleotide sequence A AAA AAG place, between orfA and the downstream fragment, orfB26,31. The slippage generates a −1 frameshift leading to a fused product of the upstream body (DNA-binding area) and the downstream body (catalytic area), ORFAB26,31. Thus, ORFAB (396 aa) from ISGmo1 is a putative protein predicted in silico32 based mostly on this mechanism. The three proteins of ISGmo1, ORFA, ORFB, and ORFAB, shared the best amino acid sequence identification with the respective open studying frames of ISFsp8 (45–49%) present in Frankia sp. and ISAar25 and ISAar4 (43–45% and 44–46%, respectively) each present in Arthrobacter arilaitensis, additionally belonging to the phylum Actinobacteria (Fig. S2). Though DDE transposases have a extremely conserved catalytic construction, they present total low sequence conservation33, which can clarify the low identities discovered between the ISGmo1 transposase and people discovered within the ISfinder database (https://www-is.biotoul.fr/). Regardless of the low identities, ISGmo1 clusters with IS sequences of different Actinobacteria, suggesting a phylogenetic relationship (Fig. S2).
In Tn6311, the inverted repeats on the left facet (IRL) consists of the beginning codon of the orfA/orfAB ISGmo1 genes and in addition two putative transposase promoters in keeping with in silico identification (http://genome2d.molgenrug.nl/) (Fig. 2a). In distinction, the repeats on the suitable facet (IRR) of ISGmo1, simply upstream the molA gene, consists of the final three codons of orfB and the − 35 area of the molA promoter. The construction of the molA gene regulatory area, i.e., the sequence of the areas − 35, − 10, and the ribosome binding sequence and their relative place in the direction of the molA begin codon (Fig. S3), suggests a robust promoter15,34. The existence of such a robust promoter mixed with the tandem group of molA gene in Tn6311 explains the excessive price of molinate breakdown by pressure ON4T, suggesting a excessive gene expression response.
Curiously, the ISGmo1 sequence was additionally present in contig A (ON4TPacBio), however with none passenger gene (Fig. 2b) and with completely different direct repeats sequences. Proof of such a duplicate of ISGmo1 was additionally given by the ON4T454, DSM13485T (JGI, AUDX00000000.1), and ON4(−) sequenced genomes and confirmed by primer design and Sanger sequencing (knowledge not proven).
Characterization of contig B: pARLON1
Contigs A and B had been presumed, based mostly on dimension, to be a chromosome and a plasmid, respectively. The detection of a band with roughly 30 kbp by Pulsed-Area Gel Electrophoresis (PFGE) (Fig. S4) supported this speculation. Furthermore, no reads mapping to contig B had been discovered within the draft genomes of strains DSM13485T (JGI, AUDX00000000.1) and ON4(−) (Fig. 1), an ON4T variant unable to degrade molinate (Fig. S5). Altogether these knowledge corroborate the existence of a cellular aspect enabling the degradation of molinate, i.e., a plasmid carrying the molA gene, which, most likely, was cured in pressure ON4(−) after 12 generations in tradition medium with out the herbicide. In flip, the absence of reads mapping to contig B in pressure DSM13485T (JGI, AUDX00000000.1) means that the situations used to develop the tradition assortment pressure didn’t favour the upkeep of the plasmid that’s fairly unstable. Therefore, contig B was named pARLON1.
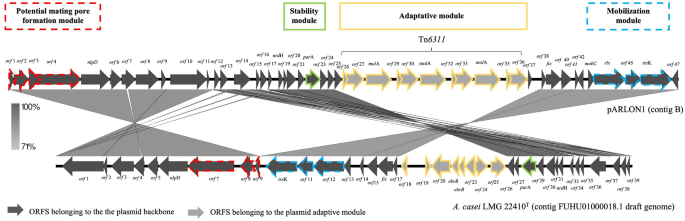
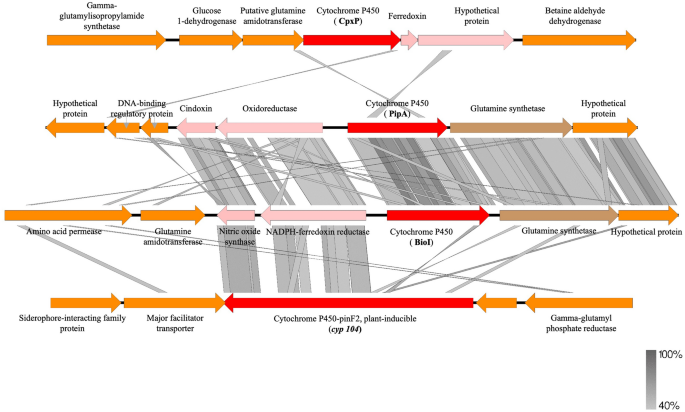
Plasmids are described as having modular constructions, the place every “module” is devoted to a specific operate (replication, stability, conjugation, institution and adaption)35. pARLON1 contained 47 recognized ORFs from which 36 ORFs (~ 27 kbp) belong to the plasmid spine, and 11 ORFs belong to the adaptive module (Tn6311, ~ 10 kbp) (Fig. 3). pARLON1 didn’t share vital nucleotide identification with different described plasmids (< 70%), not even with non-conjugative plasmids harboured by different Microbacteriaceae members (e.g., plasmids pCM1 and pCM2 of Clavibacter michiganensis subsp. michiganensis, carefully associated with G. molinativorax)36. Nevertheless, pARLON1 shared a excessive nucleotide identification (88.2%) with the contig 18 of Agrococcus casei LMG 22410T draft genome (Contig FUHU01000018.1, GenBank FUHU00000000.1), as decided by ANI37 (Desk S3). LMG 22410T is the sort pressure of Agrococcus casei, additionally a member of the household Microbacteriaceae, with a G+C content material (65 mol%) nearer to that of G. molinativorax (64 mol%) than to different Agrococcus species7,38. The pARLON1 genes sharing > 70% identification with genes of the A. casei contig had been these belonging to the plasmid spine (36 predicted genes), representing 46.3 and 53.8% of the total protection of contig B and contig FUHU01000018.1, respectively. In distinction, the anticipated genes belonging to the adaptive module of every of those genetic parts had been completely different (Fig. 3, Desk S4).
Genetic group of pARLON1 and scheme of the BLASTn search outcomes between pARLON1 (contig B) and A. casei LMG 22410T (contig FUHU01000018.1 draft genome) utilizing Easyfig software program39. Plasmid genes predicted by CONJscan40 and/or oriTDB41 as associated to a putative T4SS of the sortFATA are represented by sprint strains.
Some ORFs of each pARLON1 and contig FUHU01000018.1 are associated to the useful modules of transmissible plasmids, in keeping with the annotation of pARLON1 and the in silico predictions carried out by CONJscan40 and by oriTDB41. These analyses predicted the existence of two important proteins of the single-strand DNA conjugation equipment, the relaxase (MOB) and the sort IV coupling protein (T4CP). The previous is a essential element of the relaxosome chargeable for initiating the DNA switch in each conjugative and mobilizable plasmids, and the final {couples} the relaxase-DNA nucleoprotein complicated to the sort IV secretion system (T4SS)42. A T4SS is a big complicated of proteins spanning from the cytoplasm to the cell exterior constituting the mating-pair formation (MPF) module43,44. ORF44 and ORF12 from strains ON4T and A. casei, respectively, confirmed homology with MOB proteins, whereas ORF46/trsOk and ORF10 had been associated to the TraG/TraD household protein whose members are homologs to T4CP45,46 (Fig. 3, Desk S4). Nevertheless, solely CONJscan predicted the existence of an ORF associated to VirB4 ATPase (ORF4 and ORF7 from strains ON4T and A. casei, respectively), a marker of a T4SS presence43,44 (Fig. 3, Desk S4). Therefore, in keeping with CONJscan, each ON4T and A. casei harbour a putative T4SS of the category FATA40, which is a MPF household particular to Firmicutes, Actinobacteria, Tenericutes and Archaea43,44, agreeing with the taxonomic affiliation of each ON4T and A. casei. As a result of members of those microbial teams lack an outer membrane, a number of the core complicated T4SS parts (e.g., VirB7/10) of Agrococcus tumefaciens aren’t recognized in all of the well-characterized conjugation techniques of MPFFATA sort (e.g., pCF10 and pGO1)44. Nevertheless, proteins homologous to VirB3, VirB6 and VirB8, proteins of the inner-membrane pore, are frequent inside this method sort44. Amongst these MPF proteins, CONJscan predicted the existence of PrgH in ON4T (ORF2) and A. casei (ORF8), which present homology with VirB644. Additionally, PrgI, which is usually recommended to have homology with VirB344, was predicted in ON4T (ORF3). As well as, CONJscan predicted the presence of two different ORFs (1 and 45, and 9 and 11 from strains ON4T and A. casei, respectively) associated to accent genes detected at frequencies larger than 50% inside the MPFFATA household (FATA_prgF and FATA_cd411). Though these predictions assist the existence of a MPFFATA sort, the presence of a useful MPF module in pARLON1 must be experimentally validated.
As well as, the pARLON1 spine appears to hold a stability module. This module is especially necessary in low copy quantity plasmids, the place mechanisms, such because the partition (Par) techniques, are required to make sure the even partitioning of the DNA molecules amongst progeny (vertical transmission)35,47,48. ORF22 and ORF28 from strains ON4T and A. casei, respectively, had been predicted to be associated to the plasmid partition motor protein ParA. Nevertheless, no homologous to different proteins of this method (e.g., ParB and ParS) had been predicted. These outcomes might point out that the Par system is incomplete/not useful (which might clarify the existence of the cured strains DSM13485T and ON4(−)), or that the prevailing DNA binding proteins have low homology to recognized Par proteins (decrease homology was discovered for proteins of the ParB group than for group ParA, difficulting the classification of the previous49), or that one other upkeep mechanism is current. On the inhabitants stage, conjugative plasmids are described as low copy quantity, whereas mobilizable plasmids are usually excessive copy quantity35,50. The ratio between the plasmid (parA) and chromosomal (recA) single-copy genes in pressure ON4T was 3:1 (Desk S2), suggesting that pARLON1 is a low copy quantity plasmid51. The presence of ORFs doubtlessly related to the upkeep and conjugation modules, along with the plasmid dimension (> 30 kbp) and its low copy quantity, counsel that pARLON1 generally is a conjugative plasmid35. Nevertheless, additional research are essential to validate its conjugative exercise.
As referred to above, the adaptive module of pARLON1 differed from that of contig 18 from pressure A. casei LMG 22410T. Nevertheless, the similarity of the plasmid spine and the identification of a putative MPFFATA in strains ON4T and A. casei LMG 22410T strengthens the existence of a typical actinobacterial ancestor that, relying on the setting, acquired completely different adaptive modules. Furthermore, plasmid evolution might need been mediated by way of the exercise of the insertion sequences flanking the adaptive module, IS6100 in A. casei LMG 22410T and ISGmo1 in Tn6311 of G. molinativorax ON4T.
Identification of candidate genes concerned in ACA mineralization
Given the absence of some other catabolic gene within the neighborhood of molA, a transcriptomic strategy was used to establish the potential genes concerned in ACA mineralization. The gene expression profiles of pressure ON4T rising in molinate (mineral medium with the herbicide as the only supply of power, carbon and nitrogen, herein known as ON4TMMM) and with out molinate (management medium LB, herein known as ON4TLB) had been in comparison with establish the genes overexpressed in ON4TMMM.
After high quality management, 14,380,135 and 15,330,180 reads in every library (ON4TMMM and ON4TLB, respectively) had been retrieved. Evaluation of the reads revealed that subtractive hybridization solely eliminated a small quantity of the ribosomal RNA, since 86–90% of reads mapped to rRNA. Consequently, roughly 1.4 and 1.0 million mRNA sequence reads had been obtained from ON4TMMM and ON4TLB, respectively. After mapping the mRNA reads in opposition to the ON4T genome sequence, 95% of the 3243 anticipated transcripts had been expressed beneath the examined development situations (ON4TMMM and/or ON4TLB) (Desk S5). From the 3095 expressed transcripts, 2726 had been expressed beneath each development situations and 335 had been overexpressed in ON4TMMM in comparison with ON4TLB (Tables S6 and S7). Of those, 149 transcripts had been unique in ON4TMMM, i.e., no reads had been detected in ON4TLB. RT-qPCR validated expression ends in eight overexpressed genes in ON4TMMM. The development of expression stage was just like that of the transcriptomics experiment (Desk 2).
Metabolic pathways for Gulosibacter sp. aren’t out there in KEGG. Nonetheless, the anticipated gene merchandise from the 335 overexpressed transcripts in ON4TMMM had been mapped to the KEGG database52. Among the overexpressed predicted gene merchandise in ON4TMMM mapped to KEGG Orthology (KO) classes (Fig. S6), particularly to metabolism (e.g., amino acid, carbohydrates), environmental data processing (e.g., membrane transport) and Brite hierarchies (e.g., signaling and mobile processes), respectively. Nevertheless, no full KEGG pathway seemed to be overexpressed in ON4TMMM, particularly, none that might be associated to ACA mineralization. A lot of the predicted gene merchandise mapped to multiple metabolic pathway and/or completely different predicted gene merchandise mapped to the identical metabolic operate.
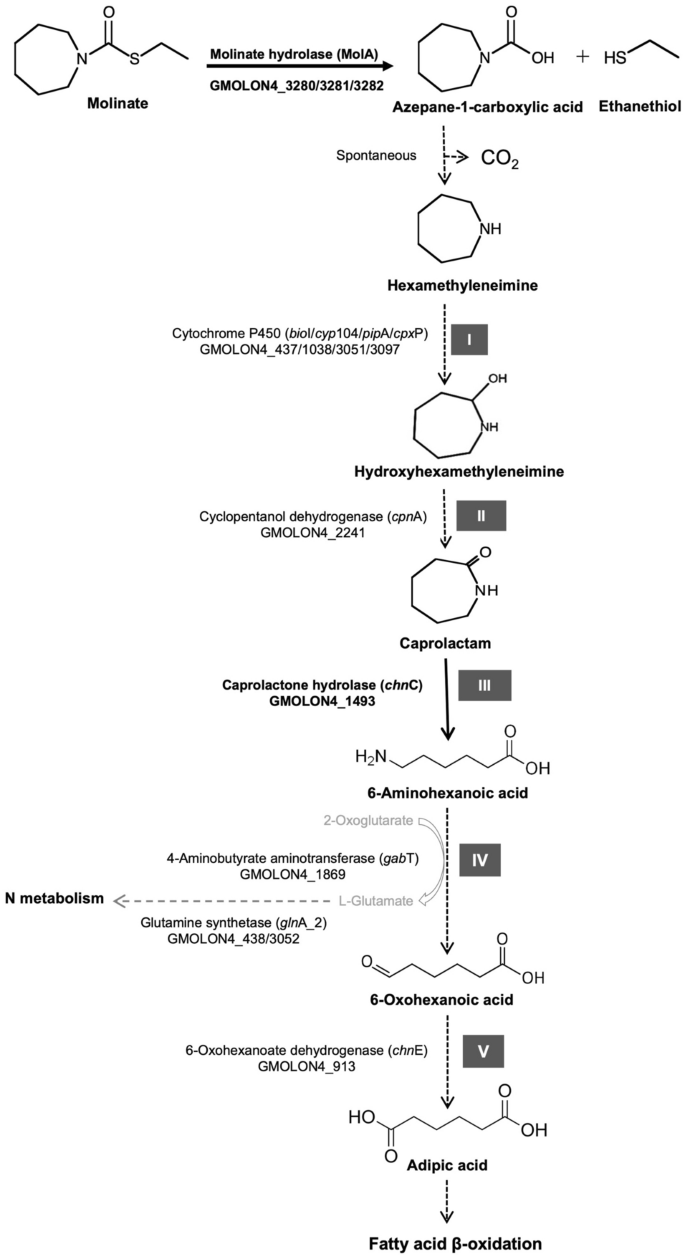
Thereby, genes doubtlessly concerned in ACA mineralization had been recognized based mostly on homology to beforehand described enzymes concerned within the degradation of different xenobiotics53,54,55,56 and on the structural similarity of the putative substrates (Fig. 4). Potential candidates concerned within the breakdown of the heterocyclic ring containing metabolites are cytochromes P450 (CYP), which act as monooxygenases and are among the many most versatile organic catalyst57, being, amongst others, concerned within the degradation of various xenobiotics57,58. In typical class I P450 techniques, the electrons required for oxygen activation are not directly supplied by NAD(P)H. The co-enzyme typically reduces a ferredoxin reductase (FDR), which in flip transfers the electrons to the second element of the system, often a ferredoxin57. Lastly, the ferredoxin mediates the electron transport between the FDR and the CYP57, additional inducing the incorporation of 1 oxygen atom into the substrate by way of the reductive cleavage of oxygen.
Candidate enzymes concerned within the 5 steps (I–V) of the putative molinate degradation pathway by pressure ON4T tailored from Barreiros and collaborators8.
Pressure ON4T harbours 4 ORFs with homology to cytochromes P450, particularly bioI, cyp104, pipA and cpxP (Fig. 5, Desk S8). Amongst these, bioI and its contiguous ferredoxin reductase (GMOLON4_437 and GMOLON4_436, respectively) had been extremely expressed in ON4TMMM (Log2FC > 6, Desk S8). Additionally, cyp104 (GMOLON4_1038) and cpxP (GMOLON4_3097) had been overexpressed in ON4TMMM (Log2FC > 3, Desk S8), whereas pipA and its contiguous ferredoxin reductase (GMOLON4_3051 and GMOLON4_3050, respectively) weren’t considerably overexpressed in ON4TMMM (Log2FC < 2, Desk S8). Noticeably, the cytochromes P450 bioI and pipA of pressure ON4T have an amino acid identification of 72% and have an analogous genetic neighbourhood (Fig. 5). Certainly, the upstream and downstream enzymes gene sequences appear to be comparable, significantly the contiguous ferredoxin reductase GMOLON4_436 and GMOLON4_3050, respectively, which have an amino acid identification of 48% with one another. Additionally, the proteins doubtlessly concerned within the switch of the electrons from the ferredoxin reductases to the cytochromes P450 bioI and pipA (GMOLON4_435 and GMOLON4_3049, respectively) share an amino acid identification of 69%. In distinction, these cytochromes (bioI and pipA) share lower than 26% and 30% amino acid identification with the cytochromes cyp104 and cpxP, respectively, that are additionally distinct from one another (24% amino acid identification) (Fig. 5).
Curiously, the cytochromes P450 bioI and pipA of pressure ON4T have an amino acid identification of roughly 53 and 52% with that of Mycobacterium smegmatis mc2155 (pipA, AAD28344.1) and Mycobacterium sp. HE5 (morA, AAV54064.1), respectively (Desk S8). Each are proven to be concerned within the hydroxylation of N-heterocycles53,59. M. smegmatis pipA was proven to be induced when pressure mc2155 grew in piperidine as the only supply of power, carbon, and nitrogen53. Mycobacterium sp. HE5 recombinant protein morA together with the adjoining pair ferredoxin reductase/ferredoxin was capable of oxidize morpholine59. Though the contiguous bioI and pipA ferredoxin reductase (GMOLON4_436 and GMOLON4_3050, respectively) had been distinct from these present in Mycobacterium strains (Fig. S7), the inhibition of ACA mineralization beneath anaerobic situations8, mixed with the similarity between the molecular construction of piperidine/morpholine (six/5 membered N-heterocycles), and the anticipated seven membered N-heterocycle product from ACA degradation in pressure ON4T (hexamethyleneimine)8 (Fig. S7), helps the involvement of one in every of these techniques within the hydroxylation of hexamethyleneimine to hydroxyhexamethyleneimine in pressure ON4T8 (step I, Fig. 4). Nevertheless, additional experimental validation is critical.
In accordance with earlier research, the ensuing hydroxyhexamethyleneimine is additional oxidized to caprolactam8, which suggests the exercise of a dehydrogenase (step II, Fig. 4). Cyclopentanol dehydrogenase (cpnA, GMOLON4_2241) is among the many overexpressed dehydrogenases in ON4TMMM (Log2FC > 2, Desk S8). It exhibits 40% amino acid identification with cyclopentanol dehydrogenases from Comamonas sp. NCIMB 987260 and 31–34% identification with cyclohexanol dehydrogenases from Rhodococcus sp. TK661 and Acinetobacter johnsonii62, respectively, recognized to oxidise cyclic alcohols however not N-heterocyclic alcohols, resembling hydroxyhexamethyleneimine63,64,65.Due to this fact, the function of the GMOLON4_2241 product within the degradation of molinate in pressure ON4T wants additional experimental validation.
Contemplating the additional step of the putative degradation of molinate, the cleavage of the cyclic metabolite (caprolactam) into 6-aminohexanoic acid (step III, Fig. 4), two potentialities arose from the transcriptomics evaluation. These had been the gene merchandise of hyuA, GMOLON4_3203, and hyuB, GMOLON4_3204 (hydantoin utilization protein A and hydantoinase B, respectively), and the gene product of chnC (GMOLON4_1493, caprolactone hydrolase). All had been overexpressed in ON4TMMM (Log2FC > 2, Desk S6). GMOLON4_3203 and GMOLON4_3204 confirmed the best amino acid identification (72–90%) with proteins of the Hydantoinase-Hydantoinase B/Oxoprolinase household discovered in numerous members of the phylum Actinobacteria. Whereas GMOLON4_1493 confirmed the best amino acid identification (75–79%) with Alpha/Beta hydrolases, adopted by caprolactone hydrolase (72–75%) of Rhodococcus sp. strains HI-31 and Phi266,67. Each hydantoinases/oxoprolinases and caprolactone hydrolase are described as capable of open cyclic natural compounds, by hydrolising C-N bonds in cyclic amides and opening the lactone ring, respectively.
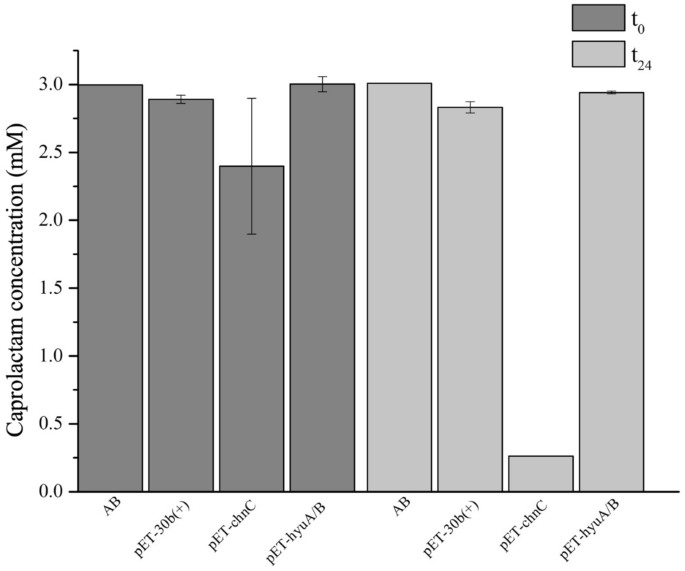
Caprolactam is the uncooked materials for manufacturing Nylon-6 by ring opening polymerization. Makes an attempt to isolate organisms able to attenuating the environmental air pollution brought on by caprolactam intensive manufacturing and utilization have been made. To date, few Gram destructive and Gram optimistic bacterial strains, together with Gulosibacter sp. BS4, had been described as able to caprolactam degradation56,68,69,70. Amongst these organisms, solely the enzyme appearing upon the cleavage of caprolactam in Pseudomonas jessenii GO3 was described56,71. This caprolactamase, which consists of two subunits with excessive sequence identification to 5-oxoprolinases, hydrolyzes caprolactam into 6-aminohexanoic acid in an ATP-dependent method56,71. Such data steered the attainable involvement of the GMOLON4_3203 and GMOLON4_3204 gene merchandise within the breakdown of caprolactam by pressure ON4T. Nevertheless, heterologous expression of each potential candidates demonstrated that solely the chnC gene product (GMOLON4_1493) degraded caprolactam in resting cells assays (Fig. 6). Proteins affiliated to this household are concerned within the cyclic ε-caprolactone transformation right into a linear natural acid, fashioned in the course of the oxidation of cyclohexanol or cyclohexanone by each Gram staining optimistic and destructive micro organism54,55,66,67. Therefore, so far as we all know, that is the primary report of their involvement in caprolactam degradation.
Analysis of caprolactam degradation in resting cells assays (DOλ600nm ≈ 6), by E. coli BL21(DE3) pressure carrying both pET-30b( +), pET-chnC or pET-hyuA/B over a 24 h incubation interval in 50 mM of phosphate buffer (pH 7.2) and three mM caprolactam. Values are means ± SE (n = 3, for all organic assays). AB—abiotic management.
Though solely 6-oxohexanoate dehydrogenase (GMOLON4_913, chnE) was overexpressed in ON4TMMM (Log2FC > 3, Desk S8), each 4-aminobutyrate aminotransferase (GMOLON4_1869, gabT) and 6-oxohexanoate dehydrogenase (GMOLON4_913, chnE) are doubtlessly concerned within the transformation of 6-aminohexanoic acid to adipic acid (step IV and V, Fig. 4). Certainly, the 4-aminobutyrate aminotransferase (GMOLON4_1869, gabT) and the 6-oxohexanoate dehydrogenase (GMOLON4_913, chnE) have an amino acid identification of 57 and 80% with the NylD1 6-aminohexanoate aminotransferase and the NylE1 adipate semialdehyde dehydrogenase from the Nylon oligomer-degrading actinobacterium Arthrobacter sp. pressure KI7224, respectively (Desk S8). In pressure KI72, NylD1 together with the cofactor pyridoxal phosphate (PLP) catalyse the transference of the amino group from 6-aminohexanoate to an amino acceptor (e.g., α-ketoglutarate, pyruvate, and glyoxylate) producing 6-oxohexanoate and an amino-acid (e.g., glutamate, alanine, and glycine), respectively. Then, NylE1 catalyses the oxidation of 6-oxohexanoate to adipate utilizing NADP+ as a cofactor, enabling the Arthrobacter sp. pressure KI72 development on 6-aminohexanoate as a sole supply of carbon and nitrogen24. Equally, the adipic acid fashioned by way of the exercise of the 6-oxohexanoate dehydrogenase (GMOLON4_913, chnE) might enter the fatty acid β-oxidation offering additional power for pressure ON4T metabolic exercise72 (Fig. 4). Certainly, the enzymes of this central metabolic pathway had been overexpressed in ON4TMMM (Desk S8). The central metabolism activated by way of succinyl-CoA and acetyl-CoA from fatty acid β-oxidation could also be thus associated to the flexibility of pressure ON4T to make use of ACA as the only supply of carbon, nitrogen, and power8.
Additionally, the overexpression of transcripts associated to the metabolism of N-compounds, significantly glutamine synthetase GMOLON4_438/3052 (Fig. 4 and Fig. S6, Desk S8), agrees with the flexibility of pressure ON4T to develop on molinate as the only supply of nitrogen8. The significance of glutamine synthetase and glutamate synthase cyclic mechanism in ammonium assimilation in prokaryotes is well-known. Particularly, Harper and collaborators73 reported that both glutamate dehydrogenase or glutamine synthase are very responsive and controlled beneath excessive or low nitrogen availability in M. smegmatis.
Among the many proteins doubtlessly concerned in molinate degradation, solely these associated with the cytochromes P450 BioI and PipA appear to be present in some strains of the genus Gulosibacter (amino acid identification of as much as 72% and 91% respectively, Desk S9). These outcomes counsel that some Gulosibacter strains, resembling G. chungangensis KCTC 13959T, might have the flexibility to partially oxidize N-heterocycle compounds.
Curiously, the merchandise of the genes putatively concerned on hexamethyleneimine degradation in pressure ON4T confirmed comparatively excessive amino acid identification with proteins concerned within the oxidation of 6 membered N-heterocycle compounds (piperidine) and cycloalkane alcohols (cyclopentanol/cyclohexanol) by a large range of strains, particularly actinobacteria (Desk S8). All these outcomes counsel that small alterations on the construction of those proteins might have allowed the utilization of caprolactam in strains ON4T and Gulosibacter sp. BS4 and ACA in pressure ON4T as power, carbon, and nitrogen sources.