A BRD9 knock-out cell-line as a software to analyze the antiviral perform of missense variants
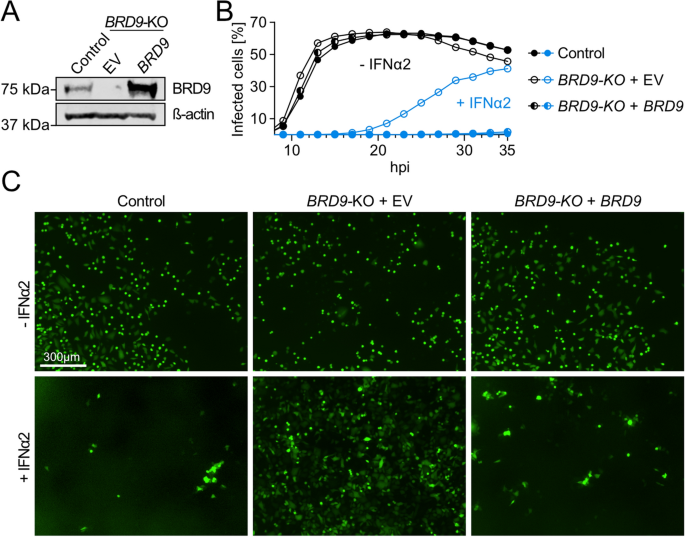
Our earlier work demonstrated that full genetic knock-out of human BRD9, or small molecule triggered degradation of the BRD9 protein, compromises environment friendly IFN-stimulated gene expression and antiviral exercise16. As an experimental software to dissect the affect of naturally occurring human BRD9 missense variants, we re-validated our beforehand described CRISPR/Cas9-generated A549-based BRD9 knockout (KO) cell-line compared to a non-edited management16. Western blot evaluation confirmed the shortage of BRD9 protein expression within the KO cell-line, and validated that BRD9 ranges might be restored by secure transduction with a lentiviral vector encoding wild-type human BRD9 (Fig. 1A). Moreover, whereas vesicular stomatitis virus (VSV-GFP; an IFN-sensitive mannequin virus) replicated equally in each non-edited management cells and BRD9-KO cells beneath baseline situations, VSV-GFP was extremely attenuated for replication in IFNα2-treated non-edited management cells, however not in BRD9-KO cells (Fig. 1B,C). That is per the precise incapacity of BRD9-KO cells to mount an environment friendly antiviral ISG response16. Importantly, BRD9-KO cells reconstituted with wild-type human BRD9 had been totally capable of mount an IFNα2-mediated antiviral ISG response in opposition to VSV-GFP (Fig. 1B,C), confirming that the defect in BRD9-KO cells is particular to lack of BRD9. These knowledge validate the BRD9-KO cell-line, with reconstitution of BRD9, as a tractable experimental system to evaluate the antiviral perform of cloned BRD9 missense variants.
Supply knowledge underlying the western blots on this determine may be discovered within the Supplementary Info file.
Characterization of a BRD9 knock-out cell-line. (A) Western blot evaluation of lysates from a non-edited management A549 cell-line and a BRD9 knock-out (KO) A549 cell-line that had been stably-transduced with lentiviruses expressing BRD9 or an empty vector (EV) management. BRD9 and β-actin had been detected with particular antibodies. (B) Cells described in (A) had been stimulated with 0 or 1,000 IU/mL of IFNα2 for 16 h previous to an infection with VSV-GFP at an MOI of 1 PFU/cell. The share of contaminated cells was quantified by way of GFP sign measurements and cell confluence over a 35 h interval. Information proven are consultant of three biologically unbiased experiments. (C) Consultant fluorescent photographs of experiments described in (B). Scale bar represents 300 μm.
Rarity of BRD9 missense variants in human genomes suggests the significance of BRD9 sequence conservation in vivo
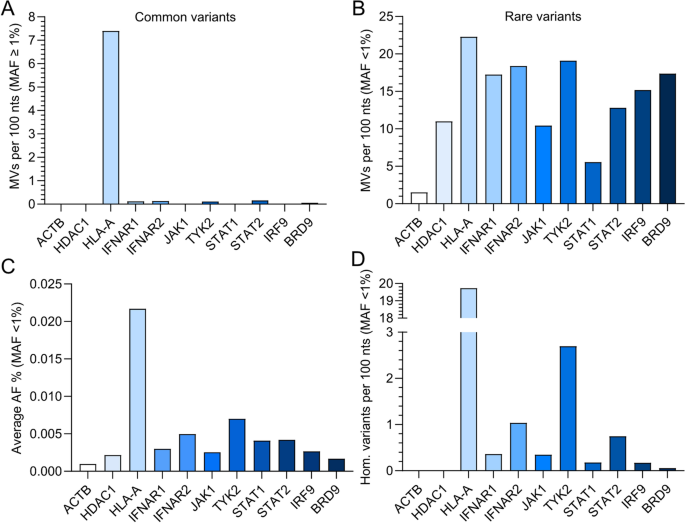
Following the same technique to others19, we analyzed all missense variants (MVs) in human BRD9 which might be contained within the gnomAD database. Thus, we in contrast the computed variability of BRD9 to the computed variabilities of canonical IFN signaling pathway element genes (IFNAR1, IFNAR2, JAK1, TYK2, STAT1, STAT2 and IRF9), recognized extremely conserved genes (ACTB and HDAC1), and a recognized polymorphic gene (HLA-A). To account for the various lengths of every gene, we calculated the variety of missense variants per 100 nucleotides (nts). Utilizing this technique, we confirmed that HLA-A is very variable, with 7.4 frequent missense variants per 100 nts (‘frequent’ outlined right here as minor allele frequency, MAF, ≥ 1%). In distinction, all different examined genes had been comparatively highly-conserved with respect to frequent missense variants, with lower than 0.2 frequent missense variants per 100 nts for IFNAR1, IFNAR2, TYK2, STAT2 and BRD9, and no frequent variants for ACTB, HDAC1, JAK1, STAT1 and IRF9 (Fig. 2A). In distinction, uncommon missense variants (‘uncommon’ outlined right here as MAF < 1%) had been related in abundance between most genes (10–20 uncommon missense variants per 100 nts), with solely STAT1 (round 5 uncommon missense variants per 100 nts) and ACTB (round 1 uncommon missense variant per 100 nts) having noticeably decrease relative numbers of uncommon missense variants (Fig. 2B). We additionally calculated the typical allele frequency (AF) for uncommon missense variants to estimate the variety of people that carry a uncommon variant, and consequently discovered that BRD9 uncommon missense variants are the second least prevalent after ACTB uncommon variants (Fig. 2C). In keeping with this, a really low variety of people had been homozygous for BRD9 uncommon missense variants (just one described within the canonical transcript: rs376958215), which is analogous to extremely conserved ACTB and HDAC1, for which no homozygous uncommon missense variant carriers have been described on gnomAD (Fig. 2D). These analyses counsel that human BRD9 is nicely conserved, and should point out that uncommon BRD9 missense variants will not be readily tolerated in people. This might be per the crucial function of BRD9 within the gene expression regulatory capabilities of the ncBAF chromatin reworking advanced20, and doubtlessly additionally the antiviral IFN system16,17,18.
Assessing genetic variation in human IFN signaling pathway elements. (A) Quantity of frequent missense variants (MVs) calculated per 100 nucleotides (nts) for the indicated human IFN signaling pathway genes as deposited to gnomAD (‘frequent’ outlined as minor allele frequency (MAF) > 1%). The conserved genes ACTB and HDAC1, in addition to the extremely variable gene HLA-A, function reference controls. (B) Quantity of uncommon missense variants (MVs) calculated per 100 nucleotides (nts) for the indicated human IFN signaling pathway genes as deposited to gnomAD (‘uncommon’ outlined as MAF < 1%). The conserved genes ACTB and HDAC1, in addition to the extremely variable gene HLA-A, function reference controls. (C) Proportion of the imply allele frequency (AF) at which uncommon genetic alleles of the indicated genes are current within the gnomAD database (MAF < 1%). (D) The variety of uncommon variants present in a homozygous method as calculated per 100 nts for every of the indicated genes (MAF < 1%).
Evaluating the affect of uncommon human BRD9 missense variants on IFN-stimulated antiviral exercise
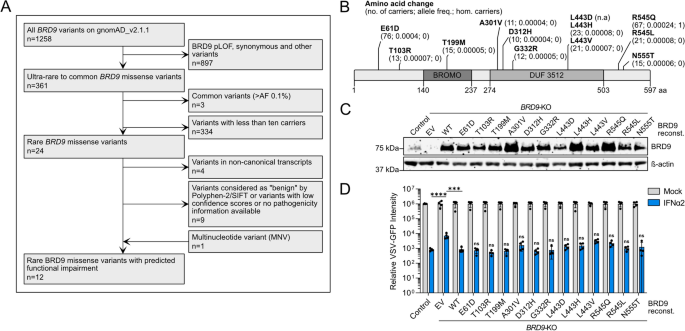
Genetic variants that harm the host, for instance by compromising the power to guard in opposition to extreme life-threatening infections, will probably be subjected to purifying choice and are thus prone to be scarce in human genomes. Nonetheless, with 1258 human BRD9 genetic variants reported on gnomAD, we initially determined to give attention to uncommon missense variants which have been described in not less than 10 people worldwide so as to set up an experimentally possible, and doubtlessly related, set of variants for useful analyses. Our filtering pipeline additionally excluded variants with synonymous mutations and mutations in splice donor/acceptor websites. Moreover, we chosen uncommon variants the place the missense mutation is predicted to affect subsequent protein performance in response to Polyphen-2 or SIFT (Fig. 3A,B and Dataset S1)21,22. The ensuing 12 BRD9 missense variants that we chosen for useful testing are distributed throughout BRD9, and a few reside in domains proven to be vital for the power of BRD9 to help IFN-mediated antiviral exercise, such because the bromodomain or the DUF3512 area (Fig. 3B)16. Utilizing site-directed mutagenesis we launched every of those variants individually right into a lentivirus-based BRD9 expression assemble, which we then used to reconstitute our BRD9-KO cell-line, thus producing isogenic cell-lines every stably expressing a unique BRD9 missense variant. As controls, the BRD9-KO cell-line was additionally reconstituted in parallel with a lentivirus vector expressing wild-type BRD9 (WT; positive-control) or an empty vector (EV; negative-control). Western blot evaluation confirmed secure expression of every BRD9 protein within the reconstituted BRD9-KO cells, and that ranges of every BRD9 missense variant had been not less than much like, or increased than, reconstituted wild-type BRD9 (Fig. 3C). To evaluate the power of every BRD9 missense variant to help IFN-mediated antiviral exercise, we subsequent pretreated cells (or not) with IFNα2 previous to an infection with VSV-GFP, and monitored GFP ranges over time as a surrogate marker for virus replication. As anticipated, IFNα2 had decreased antiviral exercise in BRD9-KO cells as in comparison with a non-edited management cell-line, and reconstitution of BRD9-KO cells with wild-type BRD9 was largely capable of reverse this phenotype. Notably, all BRD9 missense variants behaved equally to wild-type BRD9, and had been capable of restore full antiviral exercise of IFNα2 within the reconstituted BRD9-KO cells (Fig. 3D). These knowledge reveal that these chosen uncommon BRD9 missense variants, though predicted to be deleterious in nature, don’t impair the perform of BRD9 in supporting IFN-mediated antiviral exercise.
Supply knowledge underlying the western blots on this determine may be discovered within the Supplementary Info file.
Useful evaluation of uncommon human BRD9 missense variants on IFN-stimulated antiviral exercise. (A) Movement diagram mapping out the filtering pipeline used to find out uncommon human BRD9 missense variants for useful research. (B) Schematic illustration of the BRD9 area construction, with the place of missense variants highlighted. Info in brackets signifies whole variety of carriers, allele frequency, and whole variety of homozygotes. (C) Western blot evaluation of lysates from the non-edited management A549 cell-line and the BRD9 knock-out (KO) A549 cell-line stably-transduced with lentiviruses expressing every BRD9 variant, wild-type (WT) BRD9, or empty vector management (EV). BRD9 and β-actin had been detected with particular antibodies. (D) Cells described in (C) had been stimulated with 0 or 250 IU/mL of IFNα2 for 16 h previous to an infection with VSV-GFP at an MOI of 1 PFU/cell. GFP depth sign measurements taken following an infection are plotted. Information signify means and commonplace deviations from 4 biologically unbiased experiments. Particular person datapoints are proven. Statistical significance was decided utilizing a two-tailed t-test of the one time-point reworked values of IFNα2-treated situations (management vs. EV, ****p = 0.0001; EV vs. WT, ***p = 0.0002; WT vs. every indicated BRD9 variant, ns = not important).
Evaluating the affect of ultra-rare human BRD9 loss-of-function variants on IFN-stimulated antiviral exercise
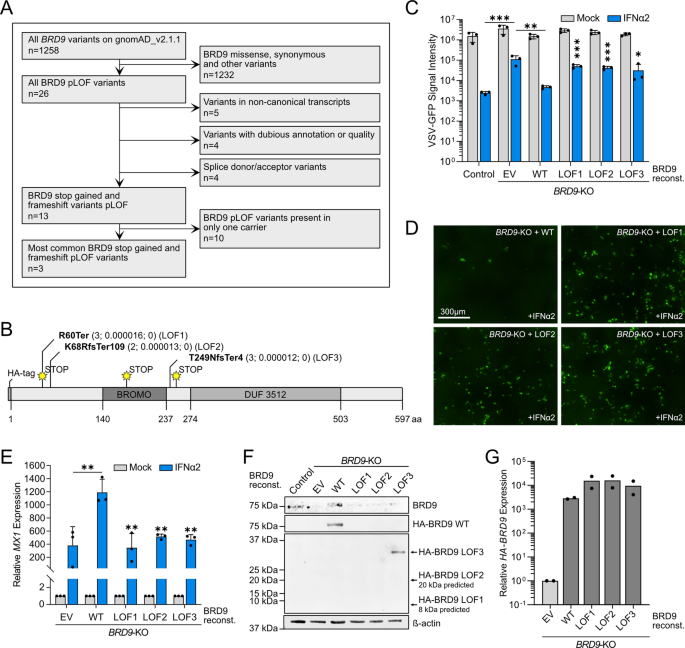
We subsequent sought to evaluate the useful affect of a lot rarer (ultra-rare) human BRD9 variants which might be predicted by the Ensembl Variant Impact Predictor23 to be loss-of-function (LOF), outlined right here as frameshift and untimely cease codon variants which might be prone to profoundly affect mRNA transcript and/or translated protein ranges. Our rationale was that rarer and extra deleterious variants might have a better observable impact on IFN-stimulated antiviral exercise as they might be extra prone to be subjected to purifying choice in the event that they had been damaging in people. Following the same filtering pipeline as above, albeit retaining variants additionally described in 2–10 people worldwide (Fig. 4A and Dataset S1), we recognized 3 naturally occurring human LOF variants in BRD9 that lead to severely truncated variations of BRD9 and which take away functionally related domains, such because the bromodomain or the DUF3512 area (Fig. 4B)16. Importantly, none of those LOF variants have been recognized as homozygous in any particular person to this point. We launched every of those ultra-rare BRD9 LOF variants individually into the lentivirus-based expression system, cloning in-frame an N-terminal HA-tag to facilitate subsequent detection. We then stably reconstituted the BRD9-KO cell-line with every of those HA-tagged BRD9 LOF variants, in addition to HA-tagged full-length wild-type BRD9 as a constructive management. Whereas IFNα2 had decreased antiviral exercise in opposition to VSV-GFP in BRD9-KO cells as in comparison with the non-edited management cell-line, reconstitution of BRD9-KO cells with HA-tagged full-length wild-type BRD9 reversed this deficit (Fig. 4C). In distinction, particular person expression of any of the three BRD9 LOF variants was unable to revive the antiviral motion of IFNα2 in opposition to VSV-GFP (Fig. 4C,D). These virus-based findings had been additional supported by gene expression evaluation of MX1 as a prototypic ISG, revealing that full-length wild-type BRD9, however not any of the three BRD9 LOF variants, permitted BRD9-KO cells to reply successfully to IFNα2 (Fig. 4E). Notably, whereas western blot evaluation might verify secure protein expression of the HA-tagged WT BRD9 and longest BRD9 LOF variant (LOF3), the shorter LOF protein variants, LOF1 and LOF2, couldn’t be detected within the reconstituted cell-lines (Fig. 4F). This failure to specific to detectable ranges is per the possible instability of such proteins with very untimely cease codons (e.g. LOF1) or lengthy stretches of nonsense (LOF2). Certainly, RT-qPCR evaluation revealed that the mRNAs encoding HA-tagged WT BRD9 and all 3 BRD9 LOF variants had been expressed to related ranges (Fig. 4G). These useful knowledge point out that severely truncated ultra-rare BRD9 LOF variants, not less than when current homozygously, are prone to render cells much less capable of mount an IFN-mediated antiviral response.
Supply knowledge underlying the western blots on this determine may be discovered within the Supplementary Info file.
Useful evaluation of ultra-rare human BRD9 loss-of-function (LOF) variants on IFN-stimulated antiviral exercise. (A) Movement diagram mapping out the filtering pipeline used to find out ultra-rare human BRD9 LOF variants for useful research. (B) Schematic illustration of the BRD9 area construction, with the place of every LOF variant highlighted. Abbreviations: fs, frameshift; Ter, termination. Info in brackets signifies whole variety of carriers, allele frequency, and whole variety of homozygotes. (C) Non-edited management cells or BRD9 knock-out (KO) cells stably-transduced with lentiviruses expressing every BRD9 LOF variant, wild-type (WT) BRD9, or empty vector management (EV) had been stimulated with 0 or 100 IU/mL of IFNα2 for 16 h previous to an infection with VSV-GFP at an MOI of 1 PFU/cell. GFP depth sign measurements taken following an infection are plotted. Information signify means and commonplace deviations from three biologically unbiased experiments. Particular person datapoints are proven. Statistical significance was decided for IFNα2-treated situations utilizing unpaired one-tailed t-tests on reworked particular time factors for every LOF variant (management vs. EV, ***p = 0.0006; EV vs. WT, **p = 0.0017; WT vs. LOF1, ***p = 0.0002; WT vs. LOF2, ***p = 0.0002; WT vs. LOF3, *p = 0.0388). (D) Consultant fluorescent photographs of IFNα2-treated situations from the experiment described in (C). Scale bar represents 300 μm. (E) RT-qPCR evaluation of MX1 mRNA within the cell-lines described in (C) with or with out pretreatment with 1000 IU/mL IFNα2 for 4 h. Values had been normalized to GAPDH mRNA ranges and are relative to the respective mock-stimulated situation. Information signify means and commonplace deviations from three biologically unbiased experiments. Statistical significance was decided for IFNα2-treated situations utilizing an unpaired one-tailed t-test (EV vs. WT, **p = 0.0002; WT vs. LOF1, **p = 0.0039; WT vs. LOF2, **p = 0.0023; WT vs. LOF3, **p = 0.0022). (F) Western blot evaluation of lysates from unstimulated cells described in (C). BRD9, the HA-tag and β-actin had been detected with particular antibodies. (G) RT-qPCR evaluation of HA-BRD9 mRNA in unstimulated cells described in (C). Values had been normalized to GAPDH mRNA ranges and are relative to the management cells. Information signify means from two biologically unbiased experiments. Particular person datapoints are proven.