We could say that the Tree of Mankind (aka Y-Haplotree) begins with “genetic Adam” (some 250,000 years in the past) and splits into progressively extra downstream branches because the timeline approaches modern-day. These downstream branches may be recognized by downstream SNP marker testing of your Y chromosome (with assessments equivalent to SNP Packs, and particularly the Large Y). This downstream Y-SNP testing helps find your place on the Tree of Mankind and probably this will show very useful for quite a lot of causes:
- It could possibly assist guarantee that you’ve got been grouped precisely in a selected “genetic household” (inside a Surname Mission, for instance)
- It could possibly assist decide your ancestral origins – at instances the precise nation, and probably even the area or county … this helps focus your genealogical analysis
- It could possibly determine your nearest genetic neighbours and their related surnames … which in flip can tie you into the family tree of a selected ‘clan’ or sept
- It could possibly determine branches inside a genetic household and which one you sit on (it will also be helpful in producing a Mutation Historical past Tree)
- It could possibly spotlight the chance of Likelihood Matches on account of Convergence amongst your record of matches
However … the Large Y check is pricey. The approach beneath tells the right way to predict the Large Y consequence with out doing the check. In that approach you’ll be able to reap the advantages of the Large Y with out truly having to do it.
The approach is known as Downstream SNP Prediction as a result of we shall be predicting what SNP markers you might be more likely to check optimistic for “downstream” i.e. approaching the fashionable period, say inside the final 500-1500 years. And the MTSA within the title stands for Matches Terminal SNP Evaluation – in different phrases, you may be analysing the terminal SNPs of every individual in your record of Y-DNA matches generated from the Y-STR check that you’ve got beforehand achieved (be it the Y-DNA-37, Y-DNA-67 or Y-DNA-111).
The approach is kind of easy. It simply takes a little bit little bit of time to finish (about 10 minutes). However there may be one main caveat – it doesn’t at all times work. And when you see the outcomes, you’ll have to make a judgement name on whether or not or not you suppose the result’s more likely to be dependable. However when it does work, it really works effectively.
Primarily the MTSA methodology entails accumulating the terminal SNPs of all your Y-STR matches after which seeing the place every SNP in flip sits on the Tree of Mankind.
If all of them sit on the identical department, then you definitely in all probability do too. In the event that they sit on extensively totally different branches, then the outcomes are untrustworthy (on this specific occasion), and the tactic has not been capable of predict which downstream SNP you might be more likely to check optimistic for. As a consequence, formal SNP testing (Large Y or in any other case) shall be needed to find out your place on the Tree of Mankind.
The Methodology
Here’s a record of the steps concerned in Downstream SNP Prediction utilizing the MTSA methodology. We’ll undergo them later intimately one after the other:
- To start out, sign up to your FTDNA account and open your Y-DNA Matches Checklist.
- Type your matches record by “Haplogroup”
- Be aware down the terminal SNPs and the way usually each happens – repeat this step for every marker stage (111, 67, 37, & 25).
- Plot every SNP in activate the Haplotree
- Assess whether or not or not the SNPs fall on a single line of descent coming down the Haplotree …
- in the event that they do, there’s a good likelihood that additionally, you will comply with this line of descent and find yourself on the identical downstream department (or a department very shut by)
- if they don’t fall on the identical single line of descent, then the approach has not labored on this occasion as a result of Convergence is current
- Make a judgement name on how dependable you suppose the outcomes are
Now let us take a look at every step intimately.
Step 1 – open your Y-DNA Matches record
Step 2 – kind your matches by Haplogroup … simply click on on the title “Y-DNA Haplogroup” and this may prepare your record of matches alphabetically by their Terminal SNP.
 |
| This particular person has 183 matches on the 25 marker stage (high left) |
Step 3 – observe down the terminal SNPs and the way usually each happens
Within the instance above, this may produce a listing like this:
- BY3441
- CTS7030
- DF13
- FGC10116
- FGC10117 (x2)
- FGC10125 (x2)
- FGC28987
- L1065
- L1335 (x8)
- and so forth …
Notes
1) I do not trouble recording the frequency of single SNPs. Thus, any SNP within the record with no quantity in brackets has solely occurred as soon as within the record.
2) I ignore any identified “upstream” SNPs (e.g. M269, L21, and so forth) as these are too far upstream to be informative.
3) this train needs to be repeated at every marker stage (111, 67, 37 & 25). In follow, the 25 marker stage seems to be essentially the most informative (at present).
Step 4 – Plot every SNP in activate the Haplotree
That is essentially the most time-consuming a part of the train however you’re going to get faster with follow. To be complete, it’s best to determine the SNP Development for every SNP in flip. The SNP Development is solely the sequence of SNPs that characterise every branching level on the road of descent to the “terminal SNP’ in query.
Thus the SNP Progressions related to the record above can be listed as follows:
- BY3441 …
- R-P312/S116 > Z290 > L21/S145 > DF13 > Z39589 > DF49/S474 > Z2980 > Z2976 > DF23 > Z2961 > FGC6540 > FGC6562 > FGC6545 > BY3442 > BY3437 > BY3441
- CTS7030 … equal to L1065
- DF13 … too far upstream
- FGC10116 … equal to FGC10117
- FGC10117 (x2) …
- R-P312/S116 > Z290 > L21/S145 > DF13 > Z39589 > L1335/S530 > L1065 > FGC10125 > FGC10117
- FGC10125 (x2) …
- R-P312/S116 > Z290 > L21/S145 > DF13 > Z39589 > L1335/S530 > L1065 > FGC10125
- FGC28987 …
- R-P312/S116 > Z290 > L21/S145 > DF13 > Z39589 > L1335/S530 > L1065 > Z16325 > S744 > S764 > FGC28987
- L1065 …
- R-P312/S116 > Z290 > L21/S145 > DF13 > Z39589 > L1335/S530 > L1065
- L1335 (x8) …
- R-P312/S116 > Z290 > L21/S145 > DF13 > Z39589 > L1335/S530
- and so forth …
Notes
1) the best method to discover the SNP Development is solely to google “YTREE” and the SNP in query. This may carry you to Alex Williamson’s Large Tree, every web page of which has the SNP Development for the actual department of the Y-Haplotree below dialogue (as within the diagram beneath for the primary SNP within the record).
2) Generally the google method will carry you to a department barely upstream of the SNP you need and you’ll have to search the webpage for the extra downstream SNP. Do that by clicking cmd+F (ctrl+F on a PC) to FIND the SNP in query.
Step 5 – Do the SNPs fall on a single line of descent?
Evaluating the SNP Progressions above, a sample clearly emerges. The vast majority of the SNP Progressions are on a single line of descent, at the least as far down as L1065. The exception is the primary SNP (BY3441), which splits off from the remainder, two branches above L1065.
Under L1065, there are at the least two branches – one through FGC10125 (5 situations – rely rigorously – rely bullet factors 4-6), the opposite through Z16325 (bullet level 7). So the SNPs do fall on a single line of descent … up to some extent. And past that time, there may be some disparity … some discordance … totally different SNPs on totally different (i.e. separate) branches of the Haplotree.
Be aware
Likelihood Matches might additionally conceivably be on account of an excessive lack of Divergence (i.e. the Y-STR signature / haplotype is handed down unchanged for a lot of hundreds of years), however the possibilities of this being the trigger are in all probability very low.
Step 6 – make a judgement name
So the place is that this specific particular person more likely to sit on the Tree of Mankind? Based mostly purely on the (partial) knowledge introduced above, he sits …
- In all probability beneath Z39589 (estimated chance … what? say … 99%? 95%?)
- In all probability beneath L1335 (estimated chance … 16 out of 17 situations = about 94%?)
- In all probability beneath L1065 (estimated chance … 8 out of 9 situations = about 89%?)
- In all probability beneath FGC10125 (estimated chance … 5 out of seven situations = about 71%?)
- In all probability beneath Z16325 (estimated chance … 1 out of seven situations = about 14%?)
- In all probability beneath DF49 (estimated chance … 1 out of 17 situations = about 6%?)
These chances are comparatively crude, however actually give a robust impression that the person in query is extremely more likely to check optimistic for L1065, and beneath that’s extra more likely to check optimistic for FGC10125 than for any of the opposite downstream SNPs.
The SNP FGC10125 seems to have arisen a while at the least 1150 years in the past, so the train has probably moved us down the Haplotree to a department that arose inside the final 1000-1500 years.
However earlier than doing any downstream SNP Pack check (the R1b-L1065 SNP Pack on this instance), it’s at all times advisable to verify that the SNP Pack truly comprises the “additional downstream” SNPs of curiosity (extracted from the record of matches’ terminal SNPs above). And on this occasion, the R1b-L1065 SNP Pack comprises all of the “extra downstream” SNPs recognized within the record above. So it will be a good selection to make on this occasion … if the person didn’t need to spend cash on the Large Y.
The Output
A number of several types of profile can emerge from this train they usually broadly fall into the next classes:
- all of the proof factors to a single downstream department of the Y-Haplotree (say, inside the final 1000 years)
- many of the proof factors to a single downstream department, however there may be some minor downstream discordance inside the final 2000 years or so, with a number of “very downstream” branches predicted
- most / all the proof factors to a main subclade department (say, about 2000-4000 years in the past) however, beneath this, many downstream branches are predicted indicating main downstream discordance
- the proof suggests a number of conflicting upstream branches of the Y-Haplotree (e.g. L21, U106, M198) and just some or not one of the proof factors to a single main subclade. Thus on this case, main upstream discordance is current and correct Downstream SNP Prediction is just not doable
Listed here are just a few examples of every profile.
Situation 1 – no discordance, all the pieces factors to a single downstream department
This situation happens with Farrell Group 2. Utilizing the MTSA methodology on a lot of this group’s members after which plotting the terminal SNPs generated onto a diagram of the Haplotree, signifies that all of them fall on a single line of descent. And predicts that the members of this group will check optimistic for the downstream SNP FGC20561.
There isn’t any or little proof that there’s Convergence on this group – all of the STR matches seem like “real” “true optimistic” matches, not one of the matches seem like Likelihood Matches on account of Convergence.
 |
| MTSA of many Farrell Group 2 members predicts they’ll check optimistic for FGC20561 |
Situation 2 – minor downstream discordance
The train described above (as an example the methodology) indicated that the person’s Y-STR matches all sat on a single line of descent as far down as Z39589. Instantly after that there was some “minor discordance” (one match examined optimistic for DF49), however the majority of the group continued downstream to L1335 and L1065. Thereafter, there was some extra discordance within the group, with 5 happening the trail of FGC10125 and one turning all the way down to Z16325. Thus, all of the proof was concordant all the way down to Z39589 (100%), a majority of the accessible proof was concordant all the way down to L1065 (89%), and a smaller majority of the accessible proof was concordant all the way down to FGC10125 (71%). And from this we are able to conclude that this particular person and his Y-STR matches share a typical ancestor on the department of the tree characterised by Z39589, and possibly share one other widespread ancestor additional downstream on the department characterised by L1065, and probably share one other widespread ancestor on the FGC10125 department.
This can be a pretty typical profile that emerges from this train. It takes you up to now down the Haplotree however no additional. Extra SNP testing shall be wanted to substantiate the predictions.
 |
| Two Discordant Downstream Branches occurring beneath main subclade R-L226 |
Situation 3 – main downstream discordance
Nonetheless, it stays clear that the person does fall beneath a significant subclade department and due to this fact the related subclade SNP Pack could also be an applicable check to take (if the person doesn’t need to buy the Large Y). The SNP Pack will should be checked to see if any related SNPs are included therein.
Within the diagram beneath, MTSA predicts that the person will sit on a department downstream of M222 (a SNP marker identified to be related to vital Convergence . Nonetheless, there are at the least 6 totally different branches beneath M222 that the MTSA methodology predicts as doable candidates for the person’s department. This individual went on to do the Large Y check and the confirmed department he truly sits on turned out to be not one of the candidates predicted by MTSA. This illustrates the significance of constructing a judgement name on the reliability of the predictions.
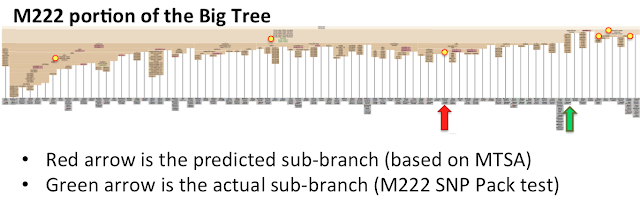 |
| A number of Discordant Downstream Branches point out main downstream discordance |
Situation 4 – main upstream discordance
Be aware
I typically use the phrases Upstream and Downstream in crude approximation to the closest main subclade, which tends to be within the vary of 2000-4000 years in the past. Upstream is roughly greater than 4000 years in the past; and Downstream is roughly lower than 2000 years in the past. However these are approximations.
Some Last Phrases
Downstream SNP Prediction utilizing the MTSA methodology may be surprisingly predictive in lots of instances.
At the moment it really works greatest on the 25 marker stage, just because there are numerous extra matches at this stage and due to this fact many extra datapoints. Nonetheless I at all times verify the upper marker ranges first and likewise verify for consistency throughout the totally different marker ranges. I’ve hardly ever explored 12 marker outcomes (as a result of the chance of Convergence at this stage is so excessive) however sometimes they will seem helpful (however, a big grain of salt wants ingestion).
Predicting the “most certainly” terminal SNP for a person permits extra focused “confirmatory” SNP testing (through a SNP Pack or single SNP check) and probably saves the shopper cash.
It additionally helps determine Likelihood Matches on account of Convergence inside a person’s match record, and thus provides some indication of the extent of Convergence inside the particular person’s match record. In subsequent weblog posts, we’ll discover how the MTSA methodology can facilitate quantification of the extent of Convergence not simply inside a person’s match record, but additionally for a complete genetic group inside a surname undertaking.
I would prefer to say a giant thanks to Ralph Taylor, James Irvine & Debbie Kennett for serving to form my concepts on this topic.
Maurice Gleeson
Dec 2017




