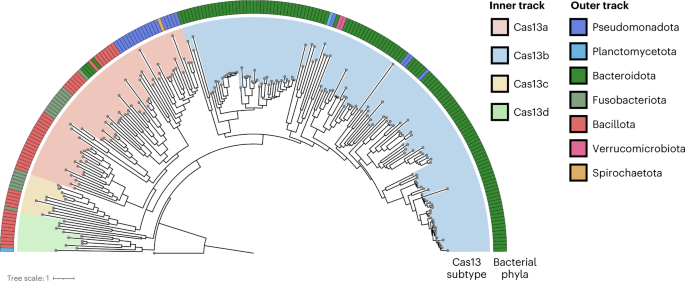
Cas13 homologues are uncommon throughout bacterial phyla
Phages encode various anti-defence methods in opposition to the bacterial defence methods they’re prone to encounter13,33,39,40, which in flip can render these methods ineffective for both phage immunity or phage engineering. To find out whether or not Cas13 is likely to be helpful as each a broad-spectrum phage defence and a phage genome enhancing software, we started by investigating the distribution of Cas13 effectors throughout bacterial phyla. We carried out a bioinformatic seek for Cas13 proteins throughout NCBI and Genome Taxonomy Database (GTDB) genomes, culminating in a non-redundant set of 224 Cas13 protein sequences (Fig. 1 and Supplementary Fig. 1). According to earlier classification efforts2, Cas13 subtypes cluster into 4 clades 13a–d. We discovered Cas13b to be most widespread, but predominantly discovered inside Bacteroidota. In distinction, Cas13c and Cas13d subtypes appeared least widespread, primarily present in Fusobacteriota and Bacillota (previously Firmicutes), respectively. We discovered Cas13a to be phylogenetically extra broadly dispersed, though comparatively restricted in whole variety of homologues, unfold throughout Pseudomonadota (beforehand Proteobacteria), Bacillota, Bacteroidota and Fusobacteriota.
The 4 identified subtypes, Cas13a–d, every type their clade (inside observe) with a skewed distribution throughout bacterial taxa (outer observe). A Vibrio cholerae Cas9 (UIO88932.1) was used because the outgroup. Cas13 subtypes and microbial taxa that encode Cas13 are denoted within the color bar.
Our outcomes are according to earlier CRISPR search endeavours, suggesting that Cas13 effectors are a number of the rarest Cas proteins at present recognized2. Though RNA-targeting type-III CRISPR-Cas methods are comparatively considerable in bacterial phyla2, we puzzled whether or not the sparse prevalence of Cas13 effectors implies that generalized resistance (for instance, by RNA recycling41) or specialised resistance (for instance, by anti-CRISPR33) to Cas13 is comparatively uncommon as effectively.
LbuCas13a is a potent anti-phage effector in opposition to phage T4
Two parsimonious explanations for the phylogenetic distribution of Cas13 effectors are that both Cas13 effectors are comparatively ineffective anti-phage methods, limiting their phylogenetic unfold owing to evolutionary stress, or that Cas13 effectors are potent anti-phage methods, however the health value of their abortive-infection-like results30,34 causes choice in opposition to cas13 loci. To discover these potentialities, we examined the anti-phage exercise of the most- and least-widely dispersed Cas13 effectors on the idea of our evaluation of bacterial phylogeny—Cas13a and Cas13d, respectively (Fig. 1). We chosen LbuCas13a from Leptotrichia buccalis and RfxCas13d from Ruminococcus flavefaciens resulting from their in depth biochemical characterization29,42,43,44,45. We moreover chosen an engineered variant of LbuCas13a (eLbuCas13a) that was lately reported to have decrease basal trans-RNA cleavage exercise and, thus, diminished toxicity when expressed in E. coli45. Notably, not one of the Cas13 orthologues right here have been investigated for anti-phage exercise. Whereas a Cas13a orthologue from Listeria seeligeri has been used to limit temperate and nucleus-forming phages19,30,33,35, LbuCas13a comes from a phylogenetically distinct sub-clade of Cas13a effectors (Supplementary Fig. 1).
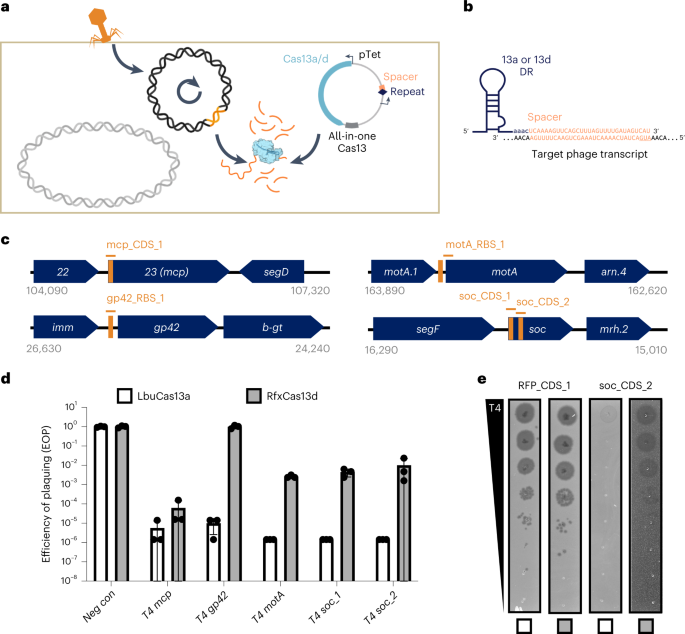
To ascertain an E. coli phage problem assay for LbuCas13a and RfxCas13d, we created ‘all-in-one’ plasmids for inducible expression of cas13 utilizing anhydrotetracycline (aTc) alongside a constitutively expressed crRNA (direct repeat-spacer) (Fig. 2a,b). Throughout phage an infection, phage RNAs are transcribed, together with a crRNA-targeted transcript (orange, Fig. 2a). Upon recognition, Cas13 prompts HEPN-mediated RNA cleavage, though the extent of trans-cleavage could also be diminished for Cas13d relative to Cas13a43. Relying on the extent of Cas13-mediated RNA cleavage, phage-encoded Cas13 resistance, protospacer mutation charge and phage-encoded operate containing the protospacer, phage could overcome the ensuing normal transcript degradation.
a, Experimental structure of Cas13 phage defence. Cas13 is expressed beneath aTc management alongside a crRNA. Throughout phage an infection, Cas13 unleashes poisonous cis– and trans-cleavage if Cas13 detects its crRNA goal. b, crRNA structure employed on this examine. c, Overview of T4 genes and transcript areas focused by Cas13 in T4 phage problem experiments. Approximate gene structure is proven in ahead orientation. crRNA areas are highlighted in orange. d, T4 phage an infection in micro organism expressing phage-targeting crRNA and both LbuCas13a or RfxCas13d. EOP values signify the common of three organic replicates for a single crRNA. EOP information are offered as imply ± s.d. e, T4 phage plaque assays evaluating the efficacy of Cas13a and toxicity of Cas13d. A consultant plaque assay from three organic replicates is proven. An RFP-targeting crRNA is proven as a unfavourable management.
To check the phage-restriction capability of LbuCas13a and RfxCas13d exterior their native context, we individually focused a small panel of genes in phage T4. Phage T4 is a classical virulent dsDNA phage with a 169 kb genome and well-characterized genetic content material46,47. From the angle of phage genome enhancing, T4 represents an empirical problem, displaying appreciable variability in Cas-restriction efficacy for Cas9 and Cas12a, owing partly to modified glucosyl-5-hydroxymethylcytosine nucleotides26,27,36 and endogenous DNA-repair mechanisms20. For these causes, we hypothesized that RNA concentrating on may very well be a superior technique to inhibit T4 and associated phages.
We designed a panel of Cas13 crRNAs concentrating on T4 transcripts with various design standards (Fig. 2c)46. Focused areas of T446 RNA sequences included important genes (main capsid protein (mcp), transcriptional activator motA), a conditionally important gene (deoxycytidylate hydroxymethylase gp42), a non-essential gene (accent capsid protein soc), an early-infection gene (motA), a middle-infection gene (gp42), late-infection genes (mcp, soc), encompassing areas early in coding sequences (CDSs) (mcp, soc), center in CDS (soc) and untranslated areas across the ribosome binding website (RBS) (gp42, motA) (Supplementary Desk 1 and Fig. 2). We included a red-fluorescent protein (RFP)-targeting crRNA as a unfavourable management. Broadly, this panel of crRNAs represents a cautious exploration of Cas13 concentrating on the variety of function varieties current in a phage transcriptome.
Remarkably, in phage an infection experiments, we noticed strong phage restriction for all crRNAs examined utilizing LbuCas13a (Fig. 2nd). Impartial of gene essentiality, timing of expression or place on transcript, we discovered that crRNA-guided LbuCas13a might prohibit phage T4 over 100,000× when concentrating on mcp, gp42, motA or soc (Supplementary Fig. 2). In distinction, crRNA-guided RfxCas13d exhibited extremely variable and less-efficient phage restriction. Additional, RfxCas13d exhibited phage-independent E. coli progress inhibition throughout RfxCas13d expression (Supplementary Figs. 2–4), and we additionally noticed a excessive diploma of phage escape for RfxCas13d relative to LbuCas13a (Fig. 2e and Supplementary Fig. 2). It’s attainable that RfxCas13d lacks essential parts required for full phage defence or diminished toxicity, such because the WYL domain-containing proteins that seem in its native gene neighbourhood (Supplementary Fig. 5). Our outcomes recommend that LbuCas13a is a remarkably potent single-protein defence system of phage T4 relative to different CRISPR-Cas methods20,26,27,36.
Cas13a confers resistance to various E. coli phages
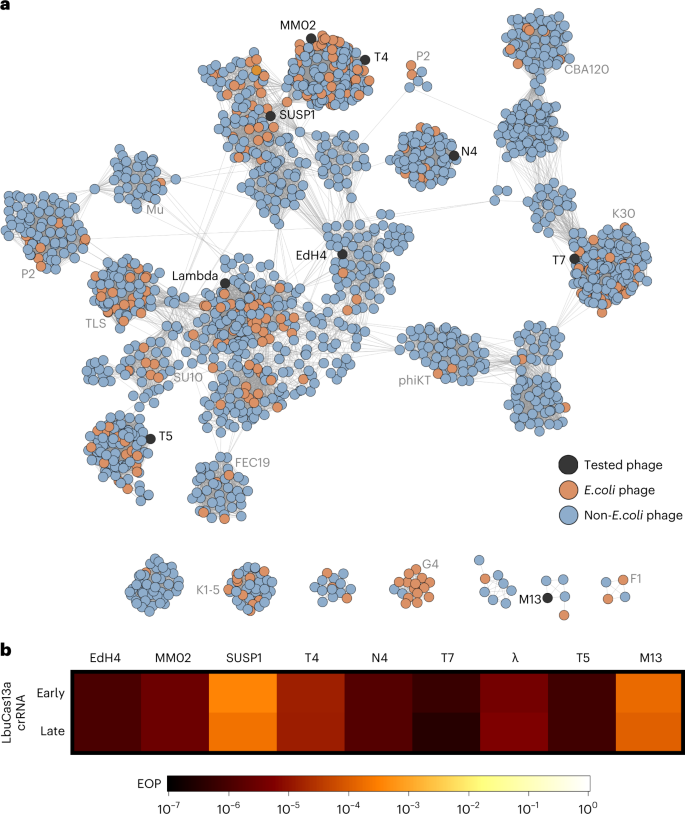
To the most effective of our information, no single Cas effector (or antiviral defence protein) has been proven to confer broad-spectrum phage resistance in opposition to various dsDNA phages. To uncover the phage phylogenetic limits of Cas13a anti-phage exercise, we challenged E. coli expressing LbuCas13a with a phylogenetically various panel of dsDNA E. coli phages. To generate a consultant sampling of E. coli phages, we constructed a protein-sharing community from 2,307 phage genomes visualizing the relatedness of at present identified E. coli phages (Fig. 3a). From this community, we assembled a panel of eight dsDNA and one single-stranded DNA (ssDNA) E. coli phages scattered throughout the E. coli phage phylogeny (Fig. 3a, Supplementary Figs. 6 and 7 and Desk 2). This panel contains each mannequin E. coli phages (T4, T5, T7, λ and M13) and non-model E. coli phages (EdH4, MM02, N4 and SUSP1). With the only exception of phages T4 and MM02, these phages bear minimal nucleotide sequence similarity to one another (Fig. 3a and Supplementary Fig. 7). Moreover, these phages have various life and mirror a sensible mannequin sampling of the genetic variety discovered amongst identified E. coli phages. One among these phages shows temperate (λ), one other shows continual an infection (M1348), whereas the remaining seven show obligately lytic life cycles. They comprise various life together with documented plasmid-transfer-promoting (that’s, ‘superspreader’)49, DNA compartmentalization16 and pseudolysogeny50 phenotypes. In mixture, these phages not solely signify genotypic variety but in addition embody a combination of host-takeover methods, modes of entry and levels of earlier characterization.
a, Community graph illustration of E. coli phages and their relations. Nodes signify phage genomes which can be related by edges in the event that they share vital similarity as decided by vContact276 (protein similarity). Nodes are shaded purple if they’re categorized as an E. coli phage and blue in the event that they solely share similarity. Nodes are shaded black in the event that they had been assessed for sensitivity to LbuCas13a. b, EOP experiments for Cas13a designed to focus on an early or late transcript. EOP values signify the common of three organic replicates for a single crRNA in comparison with an RFP-targeting unfavourable management crRNA. Phages T4, EdH4, λ, T5 and T7 have extra crRNAs that had been examined and are offered in Supplementary Figs. 2, 8, 10, 14 and 15, respectively.
For every phage, we designed a pair of Cas13a crRNAs concentrating on both a putative early gene (DNA polymerase (dnap)), RNA polymerase (rnap (T7)), a lytic regulator (cro), replication protein (II (rep) (M13)) or a putative late gene (main capsid protein (mcp VIII) (M13)). An summary of Cas13-mediated phage restriction may be present in Supplementary Desk 2, variety of crRNAs examined in Supplementary Fig. 6 and a by-phage abstract of leads to Supplementary Figs. 2, 8–15. In mixture, we noticed substantial anti-phage exercise for all 18 guides throughout the 9 phages examined (Fig. 3b and Supplementary Desk 2). Most crRNAs diminished phage infectivity 105–106-fold, with the sparse remark of mature plaque-forming models (p.f.u.). Throughout this whole examine, we noticed no mature p.f.u.s above 0.01% frequency in wildtype (wt) phage lysates (Supplementary Figs. 2, 8–15). We noticed a single information (concentrating on T5 dnap) to yield normal toxicity and progress inhibition throughout LbuCas13a induction (Supplementary Fig. 16). This constraint required us to carry out assays within the absence of induction, reaching a mere 102-fold restriction (Supplementary Fig. 14). Nevertheless, using the reduced-toxicity LbuCas13a mutant, eLbuCas13a45, we noticed each phage restriction at 106-fold (Supplementary Fig. 14) and barely diminished toxicity within the absence of phage (Supplementary Fig. 16). Thus, we imagine that the subpar phage restriction by LbuCas13a was attributed to elevated background toxicity of the T5pol spacer slightly than an incapacity to focus on this phage gene.
Apparently, SUSP1 and M13 persistently displayed a small diploma of resistance to Cas13a (Fig. 3b). Each early- and late- transcript concentrating on guides solely decreased phage infectivity 5,000–10,000-fold in contrast with all different phages exhibiting 105–106-fold infectivity discount. We additional investigated the efficacy of SUSP1-targeting crRNAs in a plate-reader assay at a variety of multiplicities of an infection (MOIs) (Supplementary Fig. 17). In comparison with a non-targeting crRNA management, we discovered that each SUSP1dnap- and SUSP1mcp-targeting guides conferred phage resistance in any respect MOIs examined, together with MOIs >10. These outcomes point out that Cas13a concentrating on not solely conferred substantial population-level safety in opposition to SUSP1 an infection, but in addition single-cell safety30. Doubtlessly, this discordance with the abortive-infection mannequin of Cas13 safety noticed beforehand30 displays a function of LbuCas13a, a function of health in a non-native host for Cas13 or a function of SUSP1 and must be investigated additional. General, we discover that LbuCas13a is able to anti-phage exercise with no recognized limits throughout the examined coliphage phylogeny.
A generalizable markerless methodology for enhancing phage genomes
The enhancing of virulent phage genomes has remained a serious problem for phage engineering and reverse genetics, largely as a result of lack of universally relevant genetic instruments or reliance on a local CRISPR-Cas system25,26,28,37,51,52,53,54. Whereas the introduction of overseas gene content material into phages is comparatively simple to carry out with homologous recombination (HR), in the end the choice or screening for these uncommon recombinants is limiting even in well-characterized phages53. On condition that LbuCas13a phage-restriction efficacy seems to have little or no variability when it comes to information (Fig. 2), goal (Figs. 2 and 3) and phage alternative (Fig. 3), we suspected that Cas13a-mediated phage restriction can be a super software for counterselection throughout phage genome enhancing. The excessive counterselection stringency noticed earlier on this examine obviates the necessity for choice markers, creating alternatives for multi-loci enhancing. Moreover, the absence of protospacer-adjacent motif (PAM) necessities for LbuCas13a concentrating on29 means that just about any place inside or close by a phage transcript may very well be edited and chosen by LbuCas13a counterselection.
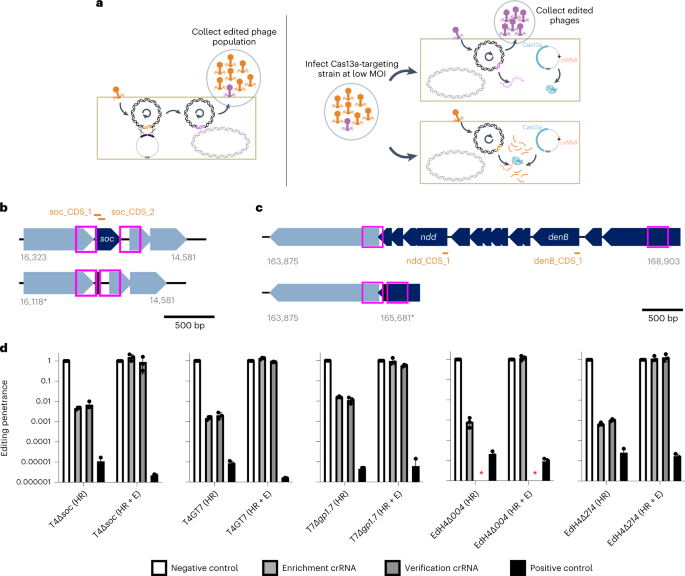
In precept, edits within the phage genome launched by homologous recombination can escape LbuCas13a concentrating on, whereas wildtype phage can’t (Fig. 4a). To introduce and choose for edits, we carried out a easy two-stage homologous recombination and enrichment course of (Fig. 4a, Supplementary Fig. 18 and Strategies). Briefly, we employed two strains per edit: an enhancing pressure containing a homologous recombination vector internet hosting a verification-primer binding website in addition to 250 bp flanking phage homology arms, and a counterselection pressure containing LbuCas13a and crRNA concentrating on the transcript carrying the locus to be edited. A pair of locus-specific examples are proven in Fig. 4b,c. We first contaminated the enhancing pressure with wildtype phage at low MOI and picked up the lysate consisting of a combination of wildtype and edited phages (‘HR’ phage lysate) (Fig. 4a and Supplementary Fig. 18a). Then we diluted this lysate, contaminated the counterselection pressure at low MOI and picked up the resultant lysate (‘HR+E’ phage lysate) (Fig. 4a and Supplementary Fig. 18b).
a, Overview of a easy two-step enhancing course of. Wildtype phage T4 infects homology vector-containing pressure at a low MOI, yielding a blended inhabitants of wt (orange) and edited (purple) phages (‘HR’). This inhabitants is diluted and infects a LbuCas13a-expressing pressure concentrating on the wt locus, enriching for edited phages relative to wt (‘HR+E’). b, Instance gene deletion design for T4∆soc. High: gene group of wt T4soc locus proven with approximate areas of soc protospacers (orange) and homology arms (pink field). Backside: gene group of edited T4∆soc locus. The encoded deletion removes each soc protospacers, enabling enrichment of edited phages. c, Instance giant multi-gene deletion design from T4gp52.1 to T4rIIB (T4wtGT7). High: gene group of wt T4GT7 locus proven with approximate areas of T4ndd and T4denB protospacers (orange) and homology arms (pink field). Backside: gene group of edited T4GT7 locus. The encoded deletion removes each soc protospacers, enabling enrichment of edited phages. d, Enhancing penetrance (Strategies) from three engineering replicates of the enhancing and enrichment course of proven in a for T4∆soc, T4GT7, T7∆gp1.7, EdH4∆gp004 and EdH4∆gp214. In all instances, ‘unfavourable management crRNA’ refers to an RFP-targeting crRNA, ‘optimistic management crRNA’ refers back to the corresponding phage’s mcp-targeting crRNA, ‘enrichment crRNA’ refers back to the crRNA used in the course of the enrichment step proven in a and ‘verification crRNA’ refers back to the deletion-targeting crRNA not used throughout enrichment. The ‘verification crRNA’ for EdH4 yielded a really poisonous phenotype to determine a titre and is denoted with a purple asterisk. Enhancing penetrance information are offered as imply ± s.d.
As a proof of idea that such an enhancing strategy is instantly relevant to reverse genetics in a variety of phages, we designed a small panel of edits throughout phages T4, T7 and EdH4. Particularly, we designed 4 single deletions (for instance, Fig. 4b) encoding for edited phages T4∆soc, T7∆gp1.7, EdH4∆gp004 and EdH4∆gp214. Whereas T4soc and T7gp1.7 (a nucleotide kinase) are identified non-essential genes46,55 beneath customary laboratory circumstances, phage EdH4 has neither been edited beforehand, neither is there any information of its genes’ essentialities earlier than this examine. Thus, EdH4 represents a stress take a look at for a way extensible this enhancing technique is to different non-model phages. For instance of extra complicated edits, we additionally designed a big edit initially recognized throughout ahead genetic screens on T4 mutant T4GT736 (hereafter, this edit within the wildtype T4 background is known as ‘T4wtGT7’). This edit consists of a giant 3.2 kb deletion in T4, totally deleting 12 genes and truncating T4gp52.1 and T4rIIB (Fig. 4c).
We designed two crRNAs disrupted by the edited phage locus of curiosity in addition to an extra verification information to verify all the gene deletion (examples for T4soc and T4wtGT7 are proven in Fig. 4b,c, respectively). When examined in opposition to wildtype phages T4, T7 and EdH4, candidate crRNAs for T4soc, T4ndd (nucleoid disruption protein), T4denB (endonuclease IV), T7gp1.7, EdH4gp004 (hypothetical protein) and EdH4gp214 (hypothetical protein) had been roughly as efficient in phage restriction as crRNAs concentrating on definitively important genes akin to mcp (Supplementary Figs. 2, 8 and 15). Nevertheless, when concentrating on these putatively non-essential genes, we noticed that plaques emerge at 10−3–10−4% frequency, doubtlessly reflecting a low charge of mutative escape permitted by the genes’ non-essentiality. In keeping with the mannequin of Cas13a primarily imparting phage defence by RNA trans-cleavage exercise, these outcomes point out that the first counterselection stress doesn’t rely on the essentiality of the crRNA goal. One among these crRNAs, the verification crRNA for the EdH4gp004 deletion, displayed elevated toxicity upon expression and is the one crRNA on this examine we couldn’t get to operate. Ostensibly, LbuCas13a’s auto-toxicity is as a result of in depth self-complementarity inside the spacer of the mature crRNA (Supplementary Fig. 19) and doubtlessly reveals a design constraint to be explored in future research.
After every stage of enhancing (Fig. 4a), lysates had been collected and titred in opposition to counterselection strains expressing LbuCas13a concentrating on the wildtype model of the edited locus (‘enrichment crRNA’ and ‘verification crRNA’), concentrating on an unedited locus (‘optimistic management crRNA’ (mcp crRNA)) and concentrating on a non-existent locus (‘unfavourable management crRNA’ (RFP crRNA)) (Supplementary Figs. 20–24). By evaluating the estimated titre in opposition to the non-targeting crRNA, we obtained a phenotypic estimate of the relative prevalence of edits inside the inhabitants (that’s, enhancing penetrance). Earlier than enrichment (‘HR’), we noticed focused Cas13a-resistant infectious centres at 0.01–1% frequency for all 5 edits (Fig. 4d). Of specific be aware, edits for EdH4 and T4wtGT7 had been typically decrease in abundance, suggesting decrease HR frequencies of enhancing for EdH4, in addition to bigger modifications. Importantly, after enrichment (‘HR+E’), focused Cas13a-resistant infectious centre edited phages comprised almost 100% of the inhabitants, whereas emergent normal Cas13 resistance remained low (Fig. 4d and Desk 1). Along with confirming edits utilizing the verification crRNA, we additionally PCR-verified 9 plaques from the ‘HR+E’ lysates noticed on the counterselection crRNA for every edit (Supplementary Figs. 25–29). Unbiased PCR-derived Sanger sequencing additional confirmed that the character of Cas13 resistance was as a result of designed edit in all instances.
PAMless Cas13a allows minimal edits in phage genomes
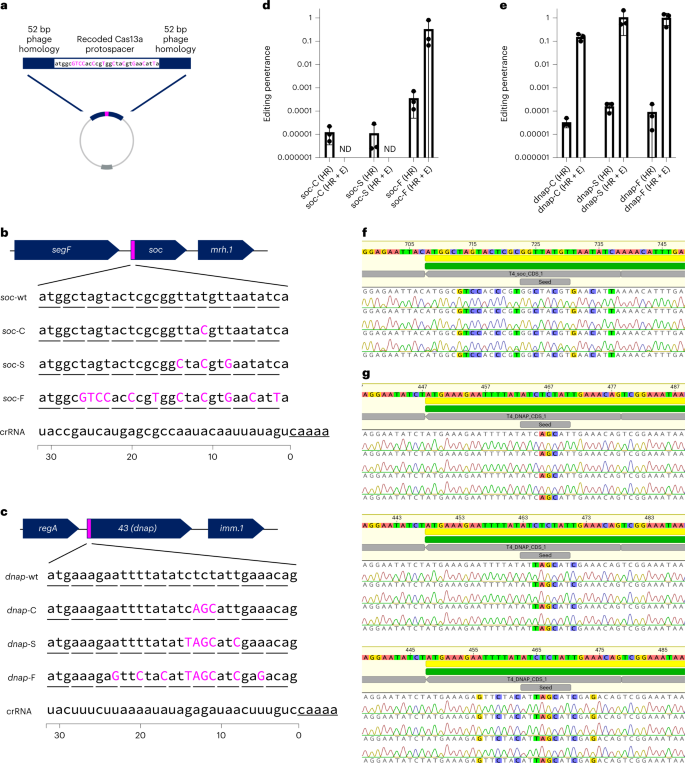
We aimed to additional make the most of the flexibleness enabled by Cas13a’s PAMless nature by creating and enriching minimal edits that solely Cas13a might simply choose for20,26,27,36, utilizing T4 as a mannequin virulent phage. We designed six mutants at both the non-essential soc gene or important dnap utilizing silent mutations, thus ‘recoding’ the goal gene (Fig. 5). We designed these mutants to recode solely a single codon (soc-C, dnap-C), recode all the seed area (soc-S, dnap-S)44 or recode the total goal (soc-F, dnap-F) (Fig. 5a–c). To facilitate homologous recombination-mediated edits, we flanked the supposed mutation with 52 bp of native phage homology (Fig. 5a). Full phenotypic outcomes from these recoding experiments for soc and dnap may be present in Supplementary Figs. 30–35.
a, Homologous recombination vector design consists of a recoded Cas13a protospacer flanked by 52 bp of homology to the phage genome. b, Recoding design for a T4 non-essential gene, soc, with launched silent mutations proven in magenta. Three designs with differing mutations had been examined (soc-C, soc-S, soc-F). Underlined nucleotides signify the sting of the Cas13a CRISPR repeat. c, Recoding design for a T4 important gene, dnap, with launched silent mutations proven in magenta. Three designs with differing levels of mutations had been examined (dnap-C, dnap-S, dnap-F). Underlined nucleotides signify the sting of the Cas13a CRISPR repeat. d, Enhancing penetrance from three organic replicates of the enhancing and enrichment course of proven in b for soc-C, soc-S and soc-F. Edited phage lysates with no detectable plaques are famous with ND. e, Enhancing penetrance from three organic replicates of the enhancing and enrichment course of proven in b for dnap-C, dnap-S and dnap-F. Enhancing penetrance in d and e are offered as imply ± s.d. f, Unbiased sequencing of T4soc loci from particular person plaques from three impartial enhancing makes an attempt. Deviations from wildtype are highlighted. g, Unbiased sequencing of T4dnap loci from particular person plaques after enhancing makes an attempt dnap-C (prime), dnap-S (center), and dnap-F (backside), every with three impartial enhancing makes an attempt. Deviations from wildtype are highlighted. Sanger sequencing traces for all verified plaques together with these proven in f and g may be present in Supplementary Figs. 16 and 20.
For 4 of the six edits (soc-F, dnap-C, dnap-S, dnap-F), we noticed that plaques emerge at 0.1–1% % frequency within the ‘HR’ lysate (Supplementary Figs. 33–35). After enrichment on the Cas13a counterselection pressure, focused Cas13a-resistant infectious centres consisted of virtually the entire phage inhabitants, suggesting excessive enhancing penetrance (Fig. 5d,e and Supplementary Figs. 33–35). In distinction, lysates containing soc-C and soc-S mutations went to extinction following enrichment, suggesting that the soc-C and soc-S mutations had been inadequate to evade Cas13a activation (Fig. 5d and Supplementary Figs. 31 and 32). Evaluating the design of soc-C, soc-S and dnap-C, a number of contiguous mutations inside the seed area seem essential to evade Cas13a activation throughout phage an infection. Doubtlessly, one of many causes we noticed only a few escape mutants in wildtype phage lysates is that a number of contiguous mutations are essential to evade Cas13a activation.
To confirm that focused Cas13a-resistant infectious centres had been the results of supposed edits, we carried out unbiased PCRs on the wildtype locus for all enhancing makes an attempt yielding plaques (Supplementary Figs. 33–35). In whole, these consisted of 36 plaques throughout 4 distinctive edits (soc-F, dnap-C, dnap-S, dnap-F) and 12 impartial enhancing processes. Strikingly, we discovered all 36 analysed plaques to have the supposed mutation (Desk 1 and Supplementary Figs. 36 and 37). It must be famous that for one engineering replicate of soc-F, we discovered two of the three plaques to encode a single nucleotide mutation (SNP) simply exterior of the location of the HR arm, in all probability reflecting an HR-induced supply of error. Nonetheless, this enhancing course of represents a easy simple route for enriching phage genome edits as small as one codon, as illustrated within the case of dnap-C.