10 years in the past we noticed a breakthrough in fashionable biology.
An American scientist found that manipulation of the Cas9 protein resulted in a gene know-how worthy of a sci-fi movie: CRISPR.
Consider it as a pair of molecular scissors able to reducing and modifying the DNA of people, animals, crops, micro organism and viruses.
The potential is large and covers something from deleting hereditary ailments to producing crops capable of stand up to local weather change.
Nevertheless, like every other new know-how, CRISPR has had its challenges. One of many primary challenges has been to make the know-how as efficient as doable and to verify the scissors solely lower the place we wish them to.
‘Now we have described new mechanisms behind CRISPR’
Two new research from the College of Copenhagen performed along with researchers from Aarhus College may help clear up these issues.
“Now we have described new mechanisms behind CRISPR,” says Professor of Bioinformatics Jan Gorodkin from the Division of Veterinary and Animal Sciences.
“We at the moment are capable of clarify why some off-targets, which is unintended cuts elsewhere within the genome, are simpler than on-targets, which is the lower on the meant place. Now we have additionally realized how totally different DNA sequences across the on-target can influence how effectively the Cas9 protein cuts the DNA. Hopefully, this data will pave the best way for simpler and safer use of CRISPR.”
So how does CRISPR work? First, a scientist will design a chunk of artificial RNA known as the information RNA. That is then connected to the Cas9 protein which can carry out the duty of reducing the DNA. The information RNA scouts for the matching DNA part. As soon as the information RNA has discovered the fitting spot, Cas9 will lower the DNA string. Now the scientist is ready to insert any artificial piece of DNA into the vacated spot.
If Cas9 and the information RNA hit the goal, the scientists discuss with it as being on course; in the event that they hit one other place, they’re off course.
At the moment, CRISPR is within the context of medication primarily used to review how genes and medicines work within the laboratory and continues to be not broadly utilized in human remedy. Nevertheless, in the long run, the thought is to make use of CRISPR within the remedy of sure genetic ailments.
Thriller of efficient off-targets solved
In one of many two new research, the researchers sought to find out one of the best ways for the information RNA to connect to the DNA, making the lower as efficient as doable—as a result of if the lower shouldn’t be sufficiently efficient, the scientists will be unable to edit the DNA.
“We already know that CRISPR does probably not work when the bond between the information RNA and the DNA is simply too weak. Now we now have realized that too sturdy a bond is problematic too,” says Gorodkin. “In each circumstances, the gene scissors are too weak and ineffective.”
The researchers then recognized an interval the place the bond between the information RNA and the DNA is neither too sturdy nor too weak, however good, leading to scissors with good sharpness.
“Curiously, this statement will also be used to clarify why some off-targets present stronger CRISPR exercise than their meant on-targets—that’s, why the scissors are sharper for some off-targets than for the on-targets,” Gorodkin explains.
“It’s because too sturdy on-targets don’t fall inside the fitting binding power interval. However when you take away a number of the power from these sturdy bonds, which is what is going on on the off-target websites, you’ll be able to get into the interval proper, leading to a extra highly effective impact and thus a shaper scissor on the off-targets.”
The examine has additionally recognized the optimum place of the Cas9 protein for reaching the best lower.
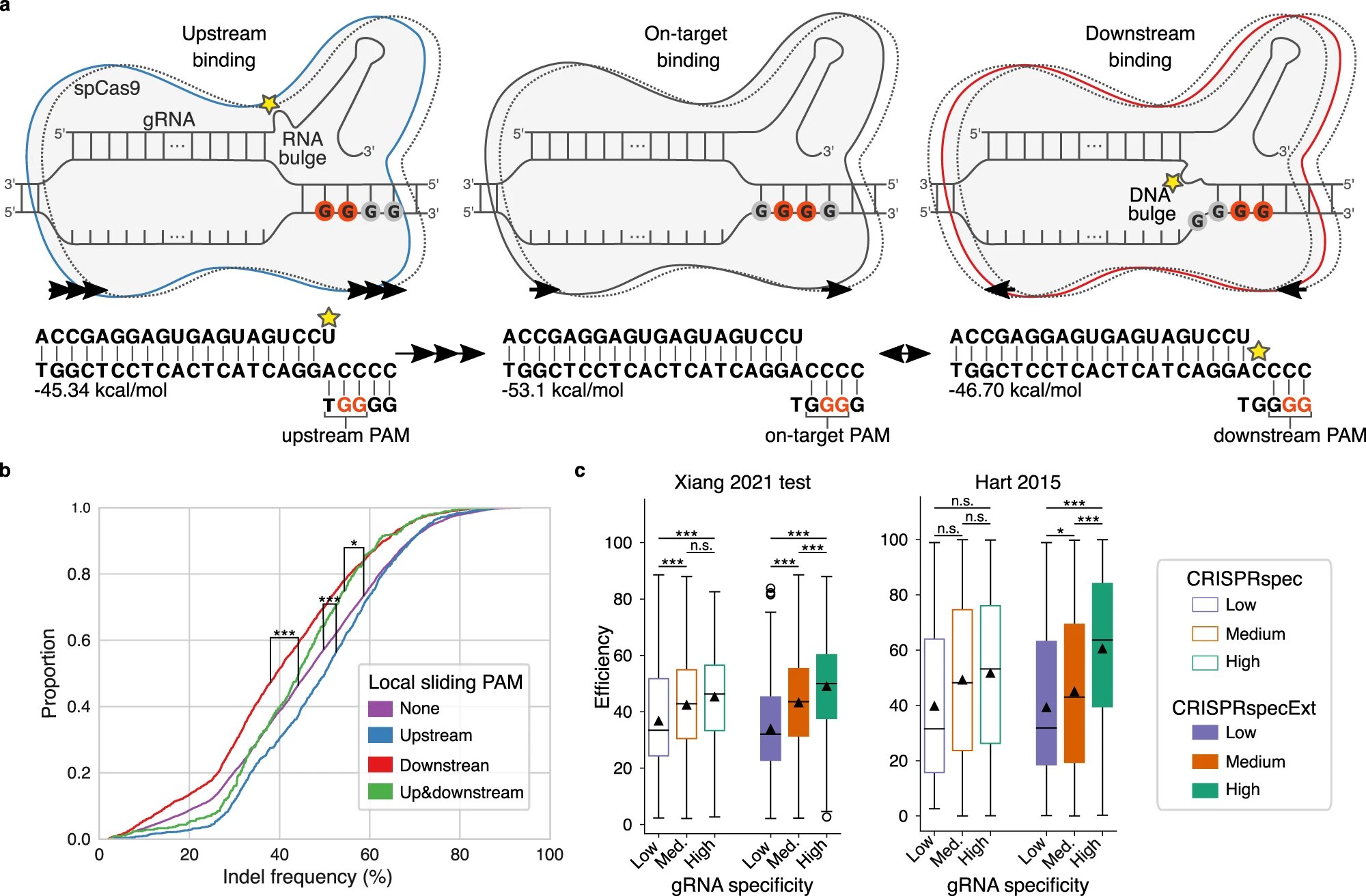
Earlier than Cas9 is ready to lower the DNA, the protein should bind to a selected a part of the DNA string. DNA consists of 4 totally different nucleotides: A, C, G and T, and Cas9 can solely bind to a sequence with two consecutive G nucleotides.
Now the researchers have recognized the impact on Cas9 of a number of consecutive G nucleotides—a scenario the place it’s laborious to hit the goal as a result of each two consecutive G’s compete for binding with Cas9.
“When there are a number of G’s ‘upstream’, that’s, earlier than the sequence that Cas9 was meant to bind to, the lower shall be simpler. However when there are a number of G’s ‘downstream’, that’s, after the meant sequence for Cas9 to bind to, the lower is much less environment friendly,” explains Postdoc Giulia Corsi.
Corsi hopes the brand new information into how CRISPR works will make it simpler to establish the right place of Cas9 sooner or later. This also needs to assist decrease the variety of potential unwanted side effects.
“We wish to have the ability to predict the lower, enhance goal modifying and get rid of off-targets, which complicate the event of recent medication by requiring numerous sources and can lead to unwanted side effects that happen while you lower the unsuitable gene,” says Corsi.
Off-targets will be dangerous—and they’re under-researched
The second examine focuses on off-targets. Right here the researchers developed a technique for measuring the effectivity of off-targets.
To quality-control a CRISPR experiment, scientists will often choose a smaller variety of computer-predicted off-targets for testing. Utilizing the brand new know-how, nevertheless, they may be capable to check a a lot bigger variety of off-targets, and that is anticipated to hurry up the event of recent medication with fewer unwanted side effects.
Utilizing the brand new methodology, the researchers examined 8,000 potential off-targets for 110 CRISPR information RNAs within the means of being translated into human medication. They discovered that round 10 % of the 8,000 potential off-targets have been in reality off-targets.
“We discovered much more off-targets than we might have been capable of utilizing current strategies,” Gorodkin explains.
Moreover, 37 of those off-targets are situated in most cancers associated genes, which will increase the danger that creating medication will develop into more durable and even not possible. As well as, unintended cuts in these genes could even result in most cancers as a doable side-effect.
“Researchers want to have the ability to establish such off-targets and choose different guide-RNAs which do not need these or every other crucial off-target,” says Gorodkin.
Nice want for extra analysis into off-targets
In line with Gorodkin, this reveals that there’s a nice want for extra analysis into off-targets.
“I might argue—and a few could disagree—that off-targets are extraordinarily under-researched. My impression is that current research on gene modifying typically lack the entire instruments and analyses required to indicate that there aren’t any off-target results of their research,” he says.
In line with Jan Gorodkin, the brand new methodology may have nice influence sooner or later to higher test research for off-targets.
“Prior to now 10 years, we now have taken a giant step in direction of having the ability to edit the genome. Now we’re within the course of of constructing our strategies higher, safer and simpler. The latter additionally helps the inexperienced transition, as genome modifications, e.g. of cells utilized in manufacturing, can result in more cost effective utilization of sources.”
The 2 research are revealed in Nature Communications.
Giulia I. Corsi et al, CRISPR/Cas9 gRNA exercise relies on free power adjustments and on the goal PAM context, Nature Communications (2022). DOI: 10.1038/s41467-022-30515-0
Xiaoguang Pan et al, Massively focused analysis of therapeutic CRISPR off-targets in cells, Nature Communications (2022). DOI: 10.1038/s41467-022-31543-6
Quotation:
Breakthrough in CRISPR analysis could result in simpler and safer gene modifying (2022, October 28)
retrieved 30 October 2022
from https://phys.org/information/2022-10-breakthrough-crispr-effective-safer-gene.html
This doc is topic to copyright. Other than any truthful dealing for the aim of personal examine or analysis, no
half could also be reproduced with out the written permission. The content material is supplied for info functions solely.