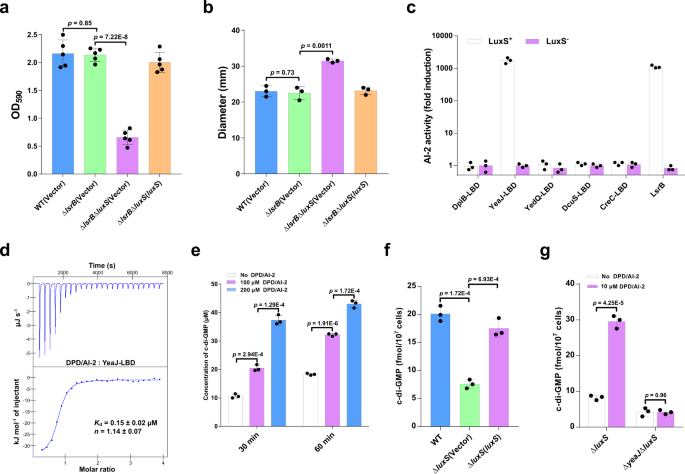
AI-2 induces c-di-GMP synthesis by appearing as an activator of YeaJ in S. Typhimurium
Though AI-2 is the well-known QS molecule produced by enteric micro organism, together with E. coli and S. Typhimurium7,26, its physiological position in these micro organism stays poorly understood. In step with a number of earlier research27,28, we discovered that deletion of luxS led to considerably decreased biofilm formation in S. Typhimurium (Supplementary Fig. 1a). Nonetheless, deletion of the lsrB gene that encodes the one recognized AI-2 receptor in S. Typhimurium26 didn’t have an effect on its skill to kind a biofilm, whereas the double mutant ΔlsrBΔluxS confirmed considerably decreased biofilm formation in contrast with the ΔlsrB mutant (Fig. 1a). Furthermore, deletion of luxS led to enhanced swimming motility in S. Typhimurium with or with out the native lsrB gene (Supplementary Fig. 1b and Fig. 1b). These outcomes recommend that AI-2 may play a job within the motile-sessile transition impartial of its receptor LsrB.
a AI-2 regulates biofilm formation in S. Typhimurium impartial of LsrB. Biofilms had been stained with crystal violet and quantified utilizing optical density measurement. Information had been imply ± s.e.m. of 5 impartial experiments. b AI-2 regulates the swimming motility of S. Typhimurium impartial of LsrB. Information had been imply ± s.e.m. of three impartial experiments. c YeaJ-LBD is able to retaining AI-2. Bioluminescence in V. harveyi MM32 (luxN−, luxS−) was induced by the addition of ligands launched from purified proteins expressed in a luxS+ or luxS– E. coli pressure. LsrB from S. Typhimurium was used as a constructive management. AI-2 exercise is reported as fold induction relative to the sunshine manufacturing induced by a buffer management. Information had been imply ± s.e.m. of three impartial experiments. d ITC assays for the particular interplay between YeaJ-LBD and AI-2. The info proven are consultant of three impartial experiments with related outcomes. The Okayd and complicated stoichiometry (n) values had been offered as imply ± s.d. of three impartial experiments. e AI-2 stimulates the DGC exercise of YeaJ in vitro. Information had been imply ± s.e.m. of three impartial experiments. f Liquid chromatography-tandem mass spectrometry (LC-MS/MS) measurements of mobile ranges of c-di-GMP in S. Typhimurium strains. The bacterial cultures grown in LB broth at 37 °C with shaking to an OD600 of 1.3 had been subjected to nucleotide extractions. Information characterize imply ± s.d. from three organic replicates. g AI-2 will increase mobile c-di-GMP focus through YeaJ. The mutants ΔluxS and ΔyeaJΔluxS grown in LB medium to an OD600 of 1.3 had been induced by 10 μM DPD/AI-2 or a buffer management for 30 min, after which bacterial cells had been collected to extract nucleotides for LC-MS/MS evaluation. Information proven are imply ± s.d. of three organic replicates. a, b, e–g Statistical significance was evaluated utilizing a two-tailed unpaired Pupil’s t-test and p < 0.05 was thought of statistically important. WT wildtype. Supply information are offered as a Supply Information file.
Given {that a} current research has discovered that dCache_1 domain-containing AI-2 receptors are extensively distributed in micro organism7, we investigated whether or not one of these AI-2 receptor is current in S. Typhimurium. Area annotations of all protein sequences of S. Typhimurium by hmmscan program in HMMER (https://www.ebi.ac.uk/Instruments/hmmer/search/hmmscan) confirmed that not one of the protein sequences has the dCache_1 area mannequin (PF02743) as the very best hit. However, periplasmic ligand-binding domains (LBDs) of 5 transmembrane proteins DpiB (STM0625), YeaJ (STM1283), YedQ (STM1987), DcuS (STM4304), and CreC (STM4589) had been discovered to hit the dCache_1 area mannequin with an E-value <1E-3. We thus examined whether or not these LBDs have the capability to bind AI-2. Within the Vibrio harveyi MM32 reporter assay, AI-2 binding exercise was noticed just for the LBD of YeaJ, however not for the LBDs of the opposite 4 proteins (Fig. 1c). Moreover, the binding evaluation by isothermal titration calorimetry (ITC) confirmed that the YeaJ-LBD binds AI-2 with a disassociation fixed (Okayd) worth of 0.15 ± 0.02 μM (Fig. 1d). These outcomes point out that AI-2 is a high-affinity ligand for YeaJ.
YeaJ has been proven to be an energetic DGC that’s concerned within the regulation of motile-sessile transition in E. coli and S. Typhimurium29,30. By the in vitro DGC exercise assay, we discovered that DPD/AI-2 stimulates the exercise of YeaJ in synthesizing c-di-GMP (Fig. 1e and Supplementary Fig. 2). In step with this discovering, when S. Typhimurium strains had been cultured to the mid-exponential part the place the extracellular AI-2 exercise reached the maximal stage within the wild-type pressure31 (Supplementary Fig. 3a), intracellular c-di-GMP stage within the ΔluxS mutant was considerably decrease than that within the wildtype (Fig. 1f). Such discount was partially restored by complementation with a plasmid encoding luxS (Fig. 1f), whereas the exogenous addition of DPD/AI-2 in cultures of ΔluxS resulted in a big improve in intracellular c-di-GMP focus (Fig. 1g). In distinction, deletion of luxS within the ΔyeaJ mutant didn’t result in important modifications within the intracellular stage of c-di-GMP (Supplementary Fig. 4), whereas the addition of DPD/AI-2 didn’t improve intracellular c-di-GMP focus in ΔyeaJΔluxS (Fig. 1g). In step with earlier research29,30, ΔyeaJ confirmed decreased biofilm formation however enhanced motility in contrast with the wildtype, whereas no important distinction in these phenotypes was noticed between ΔyeaJΔluxS and ΔyeaJ (Supplementary Fig. 5a, b), suggesting that AI-2 regulates biofilm formation and motility through YeaJ. Collectively, these outcomes point out that AI-2 positively regulates intracellular c-di-GMP ranges in S. Typhimurium by appearing as an activator of YeaJ.
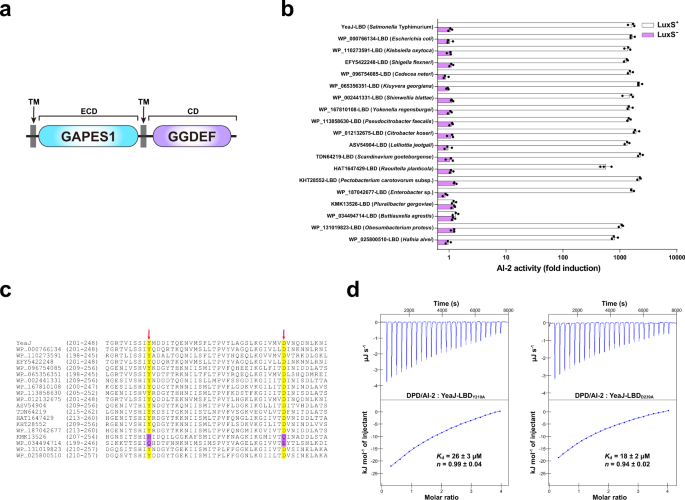
YeaJ homologs that sense AI-2 are additionally current in E. coli and different members of Enterobacterales
Whereas matching the dCache_1 area mannequin at a much less stringent E-value (1.8E-4), the LBD of YeaJ had the GAPES1 mannequin (PF17155) as the very best hit (E-value = 4.5E-153) in hmmscan searches (Fig. 2a). BLASTP looking out of the Nationwide Middle for Biotechnology Data (NCBI) non-redundant protein database adopted by area predictions utilizing InterProScan 5 in opposition to the Pfam database confirmed that YeaJ homologs that possess a putative N-terminal GAPES1 area and a putative C-terminal GGDEF area are primarily distributed within the order Enterobacterales, together with members of the households Enterobacteriaceae, Pectobacteriaceae and Hafniaceae (Supplementary Information 1 and Supplementary Fig. 6). To look at the power of the GAPES1 domains of those YeaJ homologs to bind AI-2, we randomly chosen one YeaJ homolog from every genus and ready recombinant His6-GAPES1 proteins from the luxS+ E. coli pressure. Within the V. harveyi MM32 reporter assay, AI-2 binding exercise was noticed for the GAPES1 domains of YeaJ homologs from enterohemorrhagic Escherichia coli (EHEC) O157:H7 and 15 species from different genera (Fig. 2b). In distinction, no such exercise was detected within the GAPES1 domains of two YeaJ homologs (KMK13526 and WP_034494714) from two species belonging to the genera Pluralibacter and Buttiauxella (Fig. 2b). Binding evaluation by ITC confirmed that the GAPES1 area of the YeaJ homolog from EHEC O157:H7 binds AI-2 with excessive affinity (Supplementary Fig. 7a). These information recommend that GAPES1 is a sort of extracytoplasmic sensor recognizing the AI-2 sign and YeaJ homologs whose DGC actions could be regulated by AI-2 are widespread amongst members of Enterobacterales.
a Schematic illustrating the anticipated area group of YeaJ homologs. Protein sequences had been analyzed utilizing hmmscan program in opposition to the Pfam 34.0 database. TM transmembrane area, ECD extracytoplasmic area, CD cytoplasmic area. b GAPES1 domains from YeaJ homologs in bacterial species belonging to the Enterobacteriaceae, Pectobacteriaceae, and Hafniaceae households are able to retaining AI-2. GAPES1 domains from YeaJ homologs had been expressed and purified as His6 fusion proteins in a luxS+ or luxS– E. coli pressure. Bioluminescence in V. harveyi pressure MM32 was measured following the addition of a buffer management or ligands launched from the purified proteins upon denaturing by heating. YeaJ-LBD was used as a constructive management. AI-2 exercise is offered as imply ± s.e.m. of three impartial experiments. The NCBI accession numbers for YeaJ homologs are offered and the bacterial species to which they belong are given in parentheses. c AI-2-binding GAPES1 domains of YeaJ homologs harbor two conserved residues akin to Y210 and D239 of YeaJ. GAPES1 domains of YeaJ and its homologs which were examined for AI-2 binding exercise had been aligned utilizing ClustalW embedded in MEGA7 software program. Two positions akin to Y210 and D239 of YeaJ which may be concerned in AI-2 binding are labeled with pink arrows. The conserved residues within the two positions are highlighted in yellow and non-conserved residues are highlighted in purple. The NCBI accession quantity for every YeaJ homolog is included. d Isotherms representing binding of two mutants of YeaJ-LBD (Y210A or D239A) with DPD/AI-2. The binding affinity was decided utilizing ITC. The info proven had been one consultant of three impartial experiments with related outcomes. The Okayd and complicated stoichiometry (n) are offered as imply ± s.d. of three impartial experiments. Supply information are offered as a Supply Information file.
By amino acid sequence alignment of the GAPES1 domains which were examined for AI-2 binding exercise, we discovered that two residues akin to Y210 and D239 of YeaJ are conserved in AI-2-binding GAPES1 domains however not within the two GAPES1 domains with no AI-2 binding exercise (Fig. 2c). Mutations in every of those two residues resulted in a marked discount in AI-2 binding affinity for YeaJ-LBD (Figs. 1d, 2nd). Moreover, mutations of each non-conserved residues to conserved residues inside KMK13526-LBD (H216Y/Q245D) and WP_034494714-LBD (Q208Y/E237D) elevated their AI-2 binding affinity to ranges (0.16–0.21 μM) (Supplementary Fig. 7b–e) that had been similar to that of YeaJ-LBD (Fig. 1d). These outcomes recommend that the 2 extremely conserved positions of the GAPES1 domains akin to Y210 and D239 of YeaJ could also be key residues for AI-2 binding.
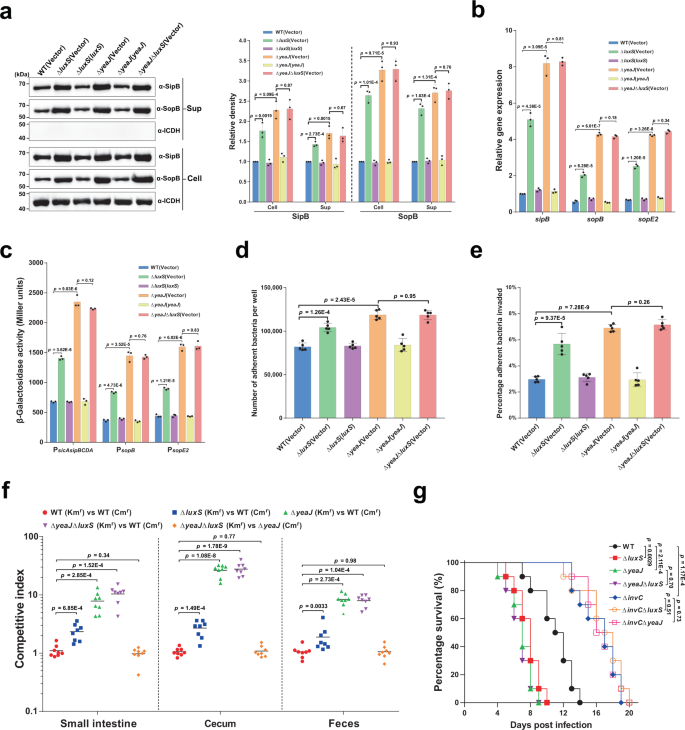
AI-2 negatively controls the T3SS-1 and attenuates the virulence of S. Typhimurium in an infection through YeaJ
Excessive ranges of c-di-GMP in S. Typhimurium have been proven to cut back the secretion of T3SS-1 effectors in addition to invasion of epithelial cells23,24,25. As anticipated, when grown underneath Salmonella pathogenicity island 1 (SPI-1) inducing situations32 to the mid-exponential part (Supplementary Fig. 3b), deletion of luxS or yeaJ considerably promoted intracellular accumulation and secretion of the T3SS-1 effectors SipB and SopB (Fig. 3a). Such induction was abolished by the expression of luxS and yeaJ within the corresponding mutants (Fig. 3a). Nonetheless, the double mutant ΔyeaJΔluxS produced and secreted SipB and SopB at ranges just like these by ΔyeaJ (Fig. 3a). These outcomes recommend that AI-2 negatively regulates the manufacturing and secretion of T3SS-1 effectors by YeaJ.
a AI-2 negatively regulates intracellular accumulation and secretion of SipB and SopB through YeaJ. Cell pellet (Cell) and concentrated supernatant (Sup) had been probed for SipB and SopB by western blot evaluation. Isocitrate dehydrogenase (ICDH) was probed as a loading management. The blots proven are consultant of three impartial experiments with related outcomes. The band intensities had been quantified by scanning densitometry utilizing ImageJ (NIH, USA), normalized to intracellular ICDH, and offered as values relative to that of the wildtype (imply ± s.d.; n = 3 impartial experiments). b mRNA ranges of T3SS-1 genes had been decided by qRT-PCR analyses. Expression was normalized to 16 S rRNA and reported as fold change relative to that of the sipB gene of the wildtype. Information had been imply ± s.e.m. of three impartial experiments. c The promoter actions of T3SS-1 genes measured utilizing β-galactosidase exercise assays (imply ± s.e.m.; n = 3 impartial experiments). d, e, Adherence to (d) and invasion of (e) Caco-2 cells by S. Typhimurium strains. Information had been imply ± s.e.m. of 5 impartial experiments. f Six-week-old feminine BALB/c mice had been contaminated orally with a 1:1 combination of two S. Typhimurium strains carrying kanamycin-resistant (Kmr) pKT100 and chloramphenicol-resistant (Cmr) pBBR1MCS1, respectively. A aggressive index (CI) was calculated because the ratio of the take a look at pressure carrying pKT100 versus (vs) the management pressure carrying pBBR1MCS1 recovered from mice. Horizontal strains characterize the geometric imply CI worth for every group (n = 8 mice per group). g Deletion of luxS or yeaJ results in enhanced virulence of S. Typhimurium in mice. Six-week-old feminine BALB/c mice had been contaminated orally with every S. Typhimurium pressure and survival was monitored each day. Information had been illustrated as a share of mice survival (n = 10 mice per group). Statistical significance was evaluated by two-tailed unpaired Pupil’s t-test (a–e), two-tailed Mann–Whitney U-test (f), or Log-rank (Mantel–Cox) take a look at (g). P values <0.05 point out important variations. Supply information are offered as a Supply Information file.
We then investigated whether or not the rise in protein ranges of SipB and SopB within the mutants ΔluxS and ΔyeaJ is because of elevated expression of sipB and sopB on the transcriptional ranges. Quantitative real-time PCR (qRT-PCR) evaluation confirmed that the mRNA ranges of sipB and sopB had been considerably greater in ΔluxS and ΔyeaJ in comparison with the wild-type and complemented strains (Fig. 3b). Related observations had been made for the expression of sopE2 (Fig. 3b), which encodes a T3SS-1 effector whose secretion can also be regulated by c-di-GMP signaling23. Furthermore, promoter-reporter assays confirmed that deletion of luxS or yeaJ led to considerably elevated promoter actions of sopB, sopE2, and the sicAsipBCDA operon (Fig. 3c). These outcomes point out that AI-2-induced elevated c-di-GMP inhibits transcription of sopB, sopE2, and the sicAsipBCDA operon.
We additional investigated the power of S. Typhimurium strains to stick to and invade human colonic epithelial Caco-2 cells. In distinction to the wild-type mum or dad pressure, mutants missing luxS or yeaJ confirmed barely enhanced adherence to (Fig. 3d), and considerably elevated invasion of Caco-2 cells (Fig. 3e). Complementation returned their adherence and invasion skill to wild-type ranges (Fig. 3d, e). Nonetheless, in distinction to our outcomes, secretion of T3SS-1 effectors and the power to invade epithelial cells weren’t altered in ΔluxS in comparison with the wildtype in a earlier research by ref. 33. We observe that totally different tradition situations had been used for each assays within the two research. In our research, each assays had been carried out utilizing cultures grown in a modified LB medium containing 0.3 M NaCl with out agitation (a situation for induction of the T3SS-1 encoded on SPI-132,34) to mid-exponential part, when the AI-2 exercise within the tradition supernatant of the wild-type pressure was maximal (Supplementary Fig. 3b), whereas the research by Perrett et al.33. used shaking cultures in regular LB medium with an OD600 of 1.0 for T3SS-1 secretion assays and within the late log part for invasion assays. We additionally discovered no variations between the wildtype and ΔluxS with respect to their skill to invade epithelial cells within the situations that ref. 33 used (Supplementary Fig. 8). Thus, the discrepancy noticed in LuxS regulation of the T3SS-1 and invasion of epithelial cells could be defined by means of totally different tradition situations.
To additional examine whether or not deletion of luxS or yeaJ impacts intestinal colonization after an infection by a pure route, we carried out aggressive oral infections of streptomycin-treated BALB/c mice with an equal combination of wild-type and mutant strains of S. Typhimurium. Competitors assays confirmed that ΔluxS, ΔyeaJ, and ΔyeaJΔluxS outcompeted the wildtype ~2 to 3-fold, ~8 to 27-fold, and ~8 to 28-fold, respectively (Fig. 3f). Aggressive indexes between ΔluxS and the wildtype, though drastically decrease than these between ΔyeaJ and the wildtype, are statistically totally different from a management competitors assay between two derivatives of the wild-type SL1344 carrying the kanamycin-resistant pKT100 and the chloramphenicol-resistant pBBR1MCS1, respectively (Fig. 3f). In distinction, when ΔyeaJΔluxS competed in opposition to ΔyeaJ, these two mutants had been recovered at related ranges within the small gut, cecum, and feces (Fig. 3f). Related outcomes had been additionally noticed when competitions had been carried out utilizing the identical strains with swapped antibiotic markers (Supplementary Fig. 9). These information recommend that AI-2 negatively regulates S. Typhimurium intestinal colonization through YeaJ.
We additionally evaluated the lethality of S. Typhimurium strains in BALB/c mice. In an oral an infection mannequin, ΔluxS and ΔyeaJ led to considerably elevated mouse mortality in comparison with the wild-type pressure, whereas infections with the mutants ΔyeaJΔluxS and ΔyeaJ produced related mortality (Fig. 3g). In step with the position of SPI-1 in intestinal an infection35,36, deletion of the gene that encodes the T3SS-1 ATPase InvC resulted in decreased mortality of mice after oral problem (Fig. 3g). Furthermore, mice contaminated with ΔinvCΔluxS and ΔinvCΔyeaJ confirmed related mortality in comparison with these contaminated with ΔinvC (Fig. 3g), indicating that AI-2-mediated c-di-GMP signaling regulates the virulence of S. Typhimurium through T3SS-1. Nonetheless, deletion of luxS or yeaJ didn’t have an effect on the virulence of S. Typhimurium after intraperitoneal inoculation (Supplementary Fig. 10), suggesting that AI-2-induced repression of the T3SS-1 through YeaJ has no main impression on systemic an infection. Collectively, these outcomes point out that AI-2 exerts a unfavourable regulatory impact on S. Typhimurium virulence throughout intestinal an infection by modulating the perform of the T3SS-1 through YeaJ.
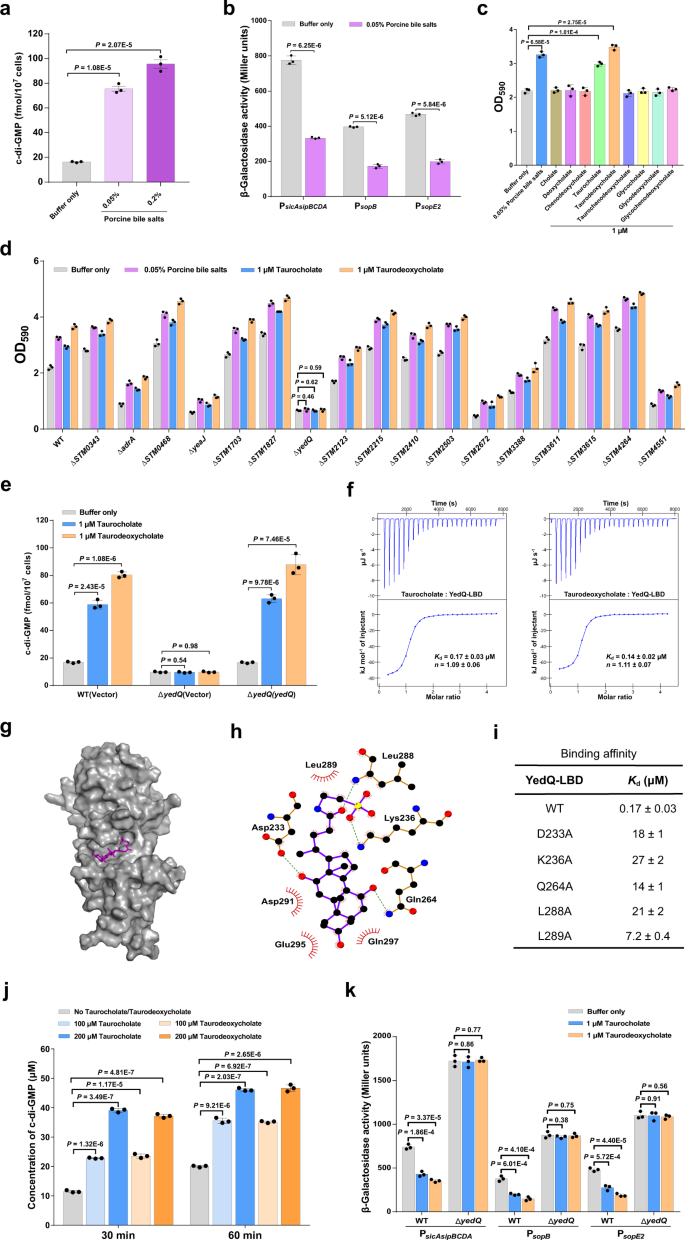
Bile salts stimulate the DGC exercise of YedQ to repress the T3SS-1 in S. Typhimurium
Bile, a serious host-produced heterogeneous combination of compounds encountered by micro organism within the small gut, was beforehand proven to repress the expression of invasion-related genes inside SPI-1 in S. Typhimurium37,38, however the mechanism of such regulation is poorly understood. Whereas bile has been reported to extend intracellular c-di-GMP ranges in V. cholerae10,14, our outcomes confirmed that AI-2 induces transcriptional repression of T3SS-1 genes through c-di-GMP signaling (Fig. 3b, c), main us to invest that bile salts could modulate intracellular c-di-GMP ranges to repress the T3SS-1 in S. Typhimurium. Certainly, when pressure SL1344 was stimulated by porcine bile salts at a focus (0.05%, w/v) similar to that physiologically occurring in intestinal contents39, the mobile focus of c-di-GMP elevated roughly fivefold (Fig. 4a). As anticipated, the promoter actions of sopB, sopE2, and sicAsipBCDA within the wild-type pressure had been considerably repressed following publicity to 0.05% bile salts (Fig. 4b). These outcomes recommend that bile salts repress T3SS-1 gene expression through growing intracellular c-di-GMP ranges in S. Typhimurium.
a Bile salts stimulate a rise in intracellular c-di-GMP concentrations. Information had been proven as imply ± s.d. of three organic replicates. b Bile salts inhibit the promoter actions of T3SS-1 genes. The promoter exercise was decided by quantifying β-galactosidase exercise. Information had been imply ± s.e.m. of three impartial experiments. c Bile salts and the person bile parts taurocholate and taurodeoxycholate stimulate biofilm formation in S. Typhimurium. Information had been imply ± s.e.m. of three impartial experiments. d Bile-induced enhancement of biofilm formation in S. Typhimurium requires YedQ. Information had been offered as imply ± s.e.m. of three impartial experiments. e A rise in intracellular c-di-GMP concentrations in response to taurocholate and taurodeoxycholate requires YedQ. Information had been imply ± s.d. of three organic replicates. f Taurocholate and taurodeoxycholate bind to the LBD of YedQ with excessive affinity. ITC information proven are one consultant of three impartial experiments with related outcomes. Okayd and complicated stoichiometry (n) are offered as imply ± s.d. of three impartial experiments. g Floor illustration of the structural mannequin of YedQ-LBD in complicated with taurocholate, ready utilizing PyMOL. Taurocholate is proven as purple sticks. h Schematic of the anticipated contacts between taurocholate and YedQ-LBD from the taurocholate-binding conformation. Potential hydrogen bonds are indicated as inexperienced dashed strains. i Binding of taurocholate to YedQ-LBD and its mutants. The binding affinity was measured by ITC. The Okayd values are offered as imply ± s.d. of three impartial experiments. j Taurocholate and taurodeoxycholate improve the DGC exercise of YedQ in vitro. Information characterize imply ± s.e.m. of three impartial experiments. Okay The promoter actions of T3SS-1 genes had been inhibited by taurocholate and taurodeoxycholate within the wild-type pressure, however not in ΔyedQ. Information had been imply ± s.e.m. of three impartial experiments. a–e, j, Okay P values had been decided utilizing the two-tailed unpaired Pupil’s t-test. A p worth lower than 0.05 was thought of to be statistically important. Supply information are offered as a Supply Information file.
In step with the recognized position of c-di-GMP in selling biofilm formation23,29, the addition of 0.05% bile salts in cultures of pressure SL1344 resulted in a big improve in biofilm manufacturing (Fig. 4c). We additional examined 9 particular person parts of bile salts to find out their contributions to biofilm formation. Intriguingly, the addition of taurocholate and taurodeoxycholate considerably stimulated biofilm formation, whereas the remaining seven parts of bile don’t have any such impact (Fig. 4c). To establish CMEs that robustly reply to bile salts, we deleted every of the 17 genes encoding predicted CMEs6 and examined the power of the mutants to kind biofilms in response to bile salts and the person bile parts taurocholate and taurodeoxycholate. Whereas most of those mutations didn’t have an effect on the response of S. Typhimurium to bile salts, the deletion of yedQ utterly abrogated the bile-induced enhancement of biofilm formation (Fig. 4d). Furthermore, the inclusion of 1 μM taurocholate or taurodeoxycholate in cultures of the wild-type pressure resulted in a big improve in intracellular c-di-GMP ranges, whereas such induction was utterly abolished in ΔyedQ (Fig. 4e). Complementation of the mutant with a plasmid derived copy of yedQ restored c-di-GMP modulation in response to taurocholate and taurodeoxycholate (Fig. 4e). These observations point out that the bile parts taurocholate and taurodeoxycholate stimulate a rise in intracellular c-di-GMP concentrations through the DGC YedQ.
We additional examined whether or not the LBD of YedQ straight interacts with taurocholate and taurodeoxycholate. Binding evaluation by ITC confirmed that taurocholate and taurodeoxycholate bind to YedQ-LBD with Okayd values of 0.17 ± 0.03 μM and 0.14 ± 0.02 μM, respectively (Fig. 4f). We then predicted the 3D construction of YedQ-LBD by Alphafold240 and carried out a docking simulation to research the interplay between YedQ-LBD and taurocholate. The very best docking conformation obtained by AutoDock Vina 1.1.241 means that taurocholate is inserted into an enormous cavity of YedQ-LBD (Fig. 4g). This conformation means that taurocholate makes shut contact with D233, K236, Q264, L288, L289, D291, E295 and Q297 within the cavity (Fig. 4h). In help of this binding mannequin, mutations in D233, K236, Q264, L288, and L289 drastically decreased the binding affinity of YedQ-LBD for taurocholate (Fig. 4i and Supplementary Fig. 11). Moreover, taurocholate and taurodeoxycholate had been capable of induce the DGC exercise of YedQ in c-di-GMP synthesis (Fig. 4j and Supplementary Fig. 12). These outcomes point out that taurocholate and taurodeoxycholate can induce intracellular accumulation of c-di-GMP in S. Typhimurium by straight partaking YedQ.
Subsequent, we examined whether or not the expression of T3SS-1 genes is affected by the presence of taurocholate and taurodeoxycholate. The addition of 1 μM taurocholate or taurodeoxycholate strongly repressed the promoter actions of sopB, sopE2, and sicAsipBCDA within the wild-type pressure, whereas this impact was utterly abolished within the ΔyedQ mutant (Fig. 4k), indicating that taurocholate and taurodeoxycholate repress T3SS-1 gene expression through YedQ. Taken collectively, our information reveal that bile parts taurocholate and taurodeoxycholate stimulate the DGC exercise of YedQ to repress T3SS-1 gene expression in S. Typhimurium.
C-di-GMP impacts the expression and secretion of T3SS-1 effectors by binding to the T3SS-1 chaperone SicA
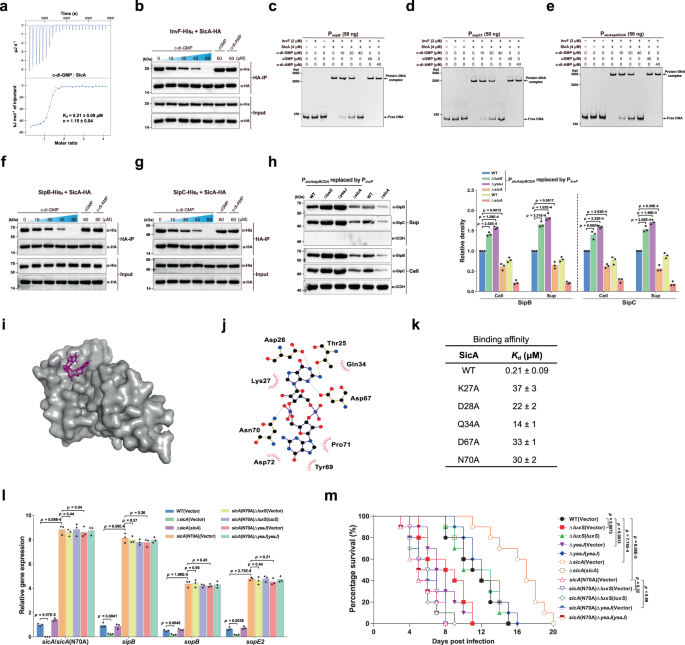
Primarily based on the above observations, we speculated that there could exist a but unidentified c-di-GMP-binding effector that regulates T3SS-1 gene expression on the transcriptional stage. It was beforehand proven that the transcription issue InvF in complicated with the T3SS-1 chaperone SicA straight prompts the expression of SipB and SopB35,42. Nonetheless, the mRNA ranges of invF in addition to its promoter exercise had been related among the many wildtype, mutants ΔluxS and ΔyeaJ, and the corresponding complemented strains (Supplementary Fig. 13), eliminating the likelihood that a rise in invF expression leads to elevated expression and manufacturing of SipB and SopB. A spread of transcription components have beforehand been proven to bind to c-di-GMP, resulting in altered DNA binding capability and, thus, modifications within the expression of downstream goal genes1,16. We thus examined whether or not InvF is a c-di-GMP effector that regulates downstream gene expression in response to this ligand. Nonetheless, ITC evaluation confirmed that InvF doesn’t bind c-di-GMP (Supplementary Fig. 14a). Of observe, plenty of small proteins had been reported to behave as c-di-GMP-binding adapters that regulate the catalytic or binding properties of their protein companions in a c-di-GMP-dependent method20,21, thus elevating one other chance that SicA is a c-di-GMP sensor. Intriguingly, binding evaluation by ITC confirmed that SicA binds c-di-GMP at a 1:1 stoichiometry with a Okayd of 0.21 ± 0.09 μM (Fig. 5a), which is similar to beforehand reported Okayd values for a number of well-established c-di-GMP receptors18,19,20,21. Against this, no binding of c-di-AMP or cGMP to SicA was detected underneath the identical experimental situations (Supplementary Fig. 14b, c). These outcomes point out that c-di-GMP straight and particularly binds to SicA.
a ITC evaluation of c-di-GMP binding to SicA. Information proven are one consultant of three impartial experiments with related outcomes, with Okayd and complicated stoichiometry (n) offered as imply ± s.d. b Co-IP of InvF-His6 with SicA-HA is impaired by c-di-GMP, however not by cGMP or c-di-AMP. The immunoprecipitated proteins (HA-IP) and the overall cell lysates (Enter) had been assessed by western blot evaluation. c–e EMSAs for InvF/SicA binding to promoters of sopB (c), sopE2 (d), and the sicAsipBCDA operon (e) within the presence and absence of nucleotides. f, g c-di-GMP decreased the Co-IP of SipB-His6 (f) and SipC-His6 (g) with SicA-HA in a dose-dependent style. h Western blot evaluation of intracellular accumulation and secretion of SipB and SipC by strains with PsicAsipBCDA changed by PinvF. Band intensities had been offered as values relative to that of the wildtype with promoter alternative (imply ± s.d.; n = 3 impartial experiments). i Floor illustration of the homology mannequin of SicA in complicated with c-di-GMP. c-di-GMP is proven as purple sticks. j Schematic of the anticipated contacts between c-di-GMP and SicA. Potential hydrogen bonds are indicated as inexperienced dashed strains. okay Binding of c-di-GMP to SicA and its mutants, as measured by ITC (Okayd = imply ± s.d.; n = 3 impartial experiments). l qRT-PCR analyses of the mRNA ranges of T3SS-1 genes. Expression was offered as values relative to that of sicA of the wildtype (imply ± s.e.m.; n = 3 impartial experiments). m Survival curves of 6-week-old feminine BALB/c mice contaminated orally with S. Typhimurium strains. Information had been illustrated as a share of mice survival (n = 10 mice per group). b–h Gels or blots proven are one consultant of three impartial experiments with related outcomes. Statistical significance was calculated utilizing the Pupil’s t-test (h, l) or Log-rank (Mantel–Cox) take a look at (m). h, l, m Variations had been thought of statistically important at p < 0.05. Supply information are offered as a Supply Information file.
We additional carried out co-immunoprecipitation (co-IP) assays to research how c-di-GMP impacts the interplay between SicA with InvF. We constructed two plasmids to specific C-terminal His6-tagged InvF and hemagglutinin (HA)-tagged SicA, respectively, and located that the co-IP of InvF-His6 with SicA-HA was impaired by c-di-GMP in a dose-dependent style, however not by excessive concentrations of c-di-AMP or cGMP (Fig. 5b). We additionally carried out electrophoretic mobility shift assays (EMSAs) to additional consider the impact of c-di-GMP on the interactions of the InvF/SicA complicated with its goal promoters. The EMSA outcomes confirmed that the InvF/SicA complicated particularly binds to the promoter sequences of sopB, sopE2, and the sicAsipBCDA operon (Supplementary Fig. 15a–c). The addition of accelerating concentrations of c-di-GMP decreased and even abrogated the formation of InvF/SicA-DNA complexes, whereas the inclusion of c-di-AMP or cGMP at a dose akin to the best stage of c-di-GMP didn’t end in an observable distinction within the InvF/SicA-DNA binding (Fig. 5c–e). Collectively, these information recommend that the binding of c-di-GMP to SicA inhibits the formation of the InvF/SicA complicated, thus leading to decreased binding of the complicated to its goal promoters.
SipB and SipC are T3SS-1 translocators that additionally act as effector proteins35,43,44. Along with appearing as a co-activator of InvF, SicA additionally capabilities to partition and stabilize SipB and SipC by binding on to them45. Immunoprecipitation experiments revealed that SicA-HA co-precipitated each SipB-His6 and SipC-His6, whereas c-di-GMP decreased the co-IP of SipB-His6 and SipC-His6 with SicA-HA in a dose-dependent style (Fig. 5f, g). We thus hypothesized that c-di-GMP may additionally have an effect on the soundness and secretion of SipB and SipC on the post-translational stage by inhibiting the binding of SicA to SipB and SipC. To experimentally take a look at this speculation, we changed the promoter of the sicAsipBCDA operon within the wild-type and mutant strains with the invF promoter, whose exercise shouldn’t be regulated by c-di-GMP signaling (Supplementary Fig. 13). After promoter alternative, the expression of sopB and sopE2 with their very own promoters was nonetheless considerably upregulated in ΔluxS and ΔyeaJ however dramatically decreased in ΔsicA when put next with the wildtype, whereas the expression of sipB and sipC underneath the management of the invF promoter was not affected by deletion of luxS, yeaJ, or sicA (Supplementary Fig. 16). In distinction, western blot evaluation confirmed that intracellular accumulation and secretion of SipB and SipC had been elevated in ΔluxS and ΔyeaJ in comparison with the wildtype, however decreased to very low ranges in ΔsicA (Fig. 5h). These outcomes help that modifications in c-di-GMP focus additionally play a job within the stability and secretion of SipB and SipC by concentrating on their chaperone SicA.
To check how SicA interacts with c-di-GMP, we constructed a homology mannequin of SicA primarily based on IpgC from Shigella flexneri (PDB ID: 3GYZ; https://www.rcsb.org/construction/3GYZ) utilizing the Phyre2 server46. Potential ligand-binding websites had been predicted utilizing POCASA 1.147 and the molecular docking was carried out by AutoDock Vina 1.1.241. The conformation with the bottom binding vitality of −8.1 kcal mol−1 (Fig. 5i) means that c-di-GMP makes shut contact with T25, K27, D28, Q34, D67, Y69, N70, P71, and D72 of SicA (Fig. 5j). Mutation of K27, D28, Q34, D67, or N70 resulted in a marked discount within the c-di-GMP-binding affinity for SicA (Fig. 5k and Supplementary Fig. 17), indicating that these residues are straight concerned in c-di-GMP binding. As well as, protein-protein docking evaluation by Cluspro 2.048 steered that InvF, SipB, and SipC have partially overlapping interplay surfaces on SicA, whereas the interplay surfaces of SicA with SipB and SipC, however not with InvF, partially overlap with the c-di-GMP-binding web site (Supplementary Fig. 18 and Fig. 5j). Along with SipB and SipC, the binding of InvF to SicA was additionally disturbed by c-di-GMP binding (Fig. 5b), suggesting that c-di-GMP allosterically regulates SicA. Amongst residues of SicA that make contact with c-di-GMP, K27, D28, Q34, and D67, however not N70, had been predicted to take part in interactions with its protein companions SipB and SipC (Supplementary Fig. 18). Certainly, the K27A variant confirmed a 19-fold decrease binding affinity to SipB in contrast with wild-type SicA (Supplementary Fig. 19a, b). In distinction, the N70A mutation of SicA didn’t have an effect on its binding affinities for InvF, SipB, and SipC (Supplementary Fig. 19a, c–g). Furthermore, altering N70 to alanine didn’t have an effect on its skill to co-immunoprecipitate InvF-His6, SipB-His6, and SipC-His6 with out the addition of c-di-GMP, whereas excessive concentrations of c-di-GMP did not impair co-IP of InvF-His6, SipB-His6, and SipC-His6 with SicAN70A-HA (Supplementary Fig. 20). These outcomes recommend that allosteric regulation of SicA by c-di-GMP is abrogated by an N70A mutation. Thus, altering N70 to alanine particularly impairs the binding of c-di-GMP however leaves its chaperone perform unaffected.
When SicAN70A was expressed at a stage just like that of wild-type SicA, the quantities of InvF, SipB, and SipC sure by SicAN70A had been a lot greater than these sure by wild-type SicA after the addition of the identical concentrations of c-di-GMP (Supplementary Fig. 20), which could be attributed to the a lot decrease c-di-GMP-binding affinity of SicAN70A and thus much less binding of SicAN70A to c-di-GMP when put next with wild-type SicA. To find out the position of c-di-GMP on SicA exercise in vivo, we changed the wild-type sicA gene within the chromosome with sicA(N70A). Whereas deletion of sicA considerably decreased the expression of sipB, sopB, and sopE2, the expression ranges of those genes had been drastically elevated within the sicA(N70A) mutant in comparison with the wild-type pressure (Fig. 5l), indicating that much less binding of SicAN70A to c-di-GMP however extra binding of this chaperone to InvF results in enhanced transcription of the goal genes of InvF/SicA within the sicA(N70A) mutant. In step with earlier findings that sicA can also be a goal gene of InvF/SicA35,42, the expression stage of the sicA(N70A) gene within the sicA(N70A) mutant was considerably greater than that of sicA within the wildtype (Fig. 5l). Moreover, deletion of luxS or yeaJ within the sicA(N70A) mutant background didn’t alter the expression of the T3SS-1 genes (Fig. 5l), suggesting that SicAN70A is unable to bind c-di-GMP at its physiological focus. Within the murine oral an infection mannequin, in distinction to the mutants ΔluxS and ΔyeaJ, the ΔsicA mutant led to considerably decreased mouse mortality in comparison with the wild-type pressure (Fig. 5m). Complementation with plasmid-borne sicA restored the lethality of the ΔsicA mutant in mice to wild-type ranges (Fig. 5m). Against this, mice contaminated with the sicA(N70A) mutant confirmed considerably greater mortality than these contaminated with the wildtype, whereas infections with sicA(N70A) and its by-product mutants missing luxS or yeaJ produced related mortality (Fig. 5m). These in vivo observations point out that the N70A mutation of the sicA gene in S. Typhimurium promotes the T3SS-1 exercise however abolishes responses to c-di-GMP signaling.
Taken collectively, these in vitro and in vivo outcomes recommend that c-di-GMP exerts its regulatory results on T3SS-1 by binding to SicA, and elevated intracellular ranges of c-di-GMP will result in much less binding of SicA to InvF, SipB, and SipC, thus downregulating the expression of the T3SS-1 genes in addition to impairing the soundness and secretion of SipB and SipC.
c-di-GMP-binding SicA homologs are extensively distributed amongst Gram-negative pathogenic micro organism
SicA of S. Typhimurium belongs to class II of T3SS chaperones and harbors three tandem tetratricopeptide repeat (TPR) motifs, which is attribute of the CesD/SycD/LcrH household of T3SS chaperones49,50,51. Homology searches utilizing the BLASTP program in opposition to the NCBI non-redundant protein database revealed that the CesD/SycD/LcrH household of chaperones that shares >20% sequence identification with SicA (E-value cutoff of 1E-03) are extensively distributed in Gram-negative micro organism, particularly the phylum Proteobacteria (Supplementary Information 2 and Supplementary Fig. 21). To look at whether or not c-di-GMP binding is a standard characteristic of this household, T3SS chaperone proteins from a number of well-known pathogenic micro organism, together with PcrH of Pseudomonas aeruginosa, IpgC of S. flexneri serotype 2a, SycD of Yersinia enterocolitica, CesD of EHEC O157:H7, BicA of Burkholderia thailandensis and VcrH of Vibrio parahaemolyticus, had been expressed and purified as recombinant proteins from E. coli BL21(DE3) and their skill to bind c-di-GMP was assessed. Strikingly, whereas these 6 chaperones share 26.2–60.1% identification with SicA, all of them had been discovered to bind c-di-GMP with excessive affinity (Okayd = 0.15–0.22 μM) (Supplementary Fig. 22). Thus, our outcomes recommend that the CesD/SycD/LcrH household of T3SS chaperones constitutes a big group of c-di-GMP effectors in Gram-negative pathogenic micro organism. Given the essential roles of the CesD/SycD/LcrH household of chaperones within the synthesis and/or secretion of T3SS effectors50,51,52, our findings help the concept that the exercise of T3SSs in a broad vary of bacterial pathogens could be modulated by c-di-GMP signaling through this household of chaperones.