Crystal construction of EsaG implies an inhibitory mechanism impartial of IDR
To analyze whether or not EsaG incorporates an IDR area as a lot of the different kind II antitoxins, we decided the crystal construction of full-length EsaG at a decision of two.30 Ǻ (Fig. 1b and Supplementary Desk 1). There are two EsaG molecules within the uneven unit, which work together with one another in a 2-fold symmetry. To analyze the oligomer state of EsaG in answer, we carried out gel-filtration experiments, chemical cross-linking assay and density gradient centrifugation evaluation. Despite the fact that gel-filtration experiments indicated a dimer state (Supplementary Fig. 1a), chemical cross-linking of EsaG utilizing an amine-specific crosslinking reagent EGS (ethylene glycol bis(succinimidyl succinate)) yielded no cross-linked product (Supplementary Fig. 1b). Density gradient centrifugation evaluation additionally supported a monomer type (Supplementary Fig. 1c). The construction of EsaG incorporates a precept four-stranded β-sheet sandwiched by αA’, αB’ and αC’ from one aspect and αD’ and αE’ from the opposite aspect (Fig. 1b). Structural comparability of EsaG with different protein buildings utilizing the Dali server25 indicated a big homology with the construction of BH3703 (PDB code 3I0T), with a Z-score of 18.9 and a root-mean-square deviation (RMSD) of 1.9 Å (Supplementary Fig. 2). BH3703 can be a putative antitoxin produced by the Gram-positive bacterium Alkalihalobacillus okuhidensis, however its organic perform stays unknown. We seen that, besides for 3 residues (G50–S51 on loopβ2’-αB’ and E162 on the C-terminus), electron densities had been noticed for all the opposite residues of EsaG. Equally, BH3703 can be effectively structured with solely three residues within the C-terminus lacking. This means the absence of IDRs in EsaG-like antitoxins and, subsequently, a definite toxin inhibition mechanism in Gram-positive micro organism.
Important conformational modifications on EsaD upon EsaG binding
To analyze the mechanism by which EsaG inhibits EsaD, we subsequent solved the crystal buildings of EsaD and the EsaD-EsaG advanced. For EsaD, we centered on its nuclease area, as a result of it has been reported that the interplay of EsaG with the nuclease area is ample to neutralize the toxicity of EsaD19. Resulting from cytotoxicity, the EsaD proteins used for crystallization had been over-expressed in E. coli in an inactive state by mutating the catalytic H528 residue to Alanine (H528A). The crystal buildings had been decided at resolutions of two.30 Å for the nuclease area of EsaD (residues 440–614) and a couple of.60 Å for the EsaG-EsaD (residues 450–614, hereafter referred as EsaDc) advanced, respectively (Fig. 1c, d and Supplementary Desk 1).
The nuclease area of EsaD presents a typical fold of ββα-metal nucleases (often known as His-Me finger nucleases)26, regardless that they share low sequence similarity (<20%). The construction incorporates a central 6-stranded β-sheet that’s largely open to solvent on one aspect and flanked by one brief α-helix (αA) and the well-known ββα-metal finger motif on the opposite aspect (Fig. 1c). The ββα-metal finger (Residues 501–573), enjoying a vital function in nicking DNA, creates a compact catalytic heart that’s composed of two antiparallel β-strands (β4 and β5) linked by an extended loop and linked to a C-terminal α-helix (αB). As noticed in another His-Me finger nucleases, αB is interrupted by a protruding ‘finger loop’ of unknown perform27. Buried inside the catalytic web site, a hydrated Mg2+ ion is immediately coordinated by a extremely conserved Asn residue (N551) positioned in αB28,29,30. As well as, an extended N-terminal loop (Residues 455–468) wraps across the central β-sheet from the β1 aspect and extends to the “again” face.
Apart from refined actions in some loops, no apparent conformational change occurs on EsaG upon EsaDc binding (Supplementary Fig. 3), additional supporting the absence of IDR within the inhibition course of. Nonetheless, the toxin, EsaD, undergoes important conformational modifications throughout the advanced formation. The central 6-stranded β-sheet of EsaDc is cut up into two impartial fragments (Fig. 1d–f): the N-terminal fragment (NF) consists of β1-3, which merges with the central β-sheet of EsaG by β4’ of EsaG and β3 of EsaDc; Within the C-terminal fragment (CF), αB from the ββα-metal finger and β6-8 work together with EsaG from the opposite aspect of the central β-sheet of EsaG. We couldn’t hint the Mg2+ ion within the catalytic heart, the lengthy loop flanking the N-terminus of NF (Residues 450–468) and a part of the ββα-metal finger motif, together with αA to the finger loop (Residues 504–556). Noticeably, the N-terminus and C-terminus of the lacking ββα-metal finger motif find on two reverse sides of EsaG, with a linear distance of about 44 Å (Supplementary Fig. 4). These observations indicated that the separation of NF and CF of EsaDc by EsaG could stretch the initially folded ββα-metal finger motif to make it turn out to be a disordered loop and stride throughout such an extended distance, thus abolishing the DNase exercise of EsaDc.
The interactions between EsaDc and EsaG
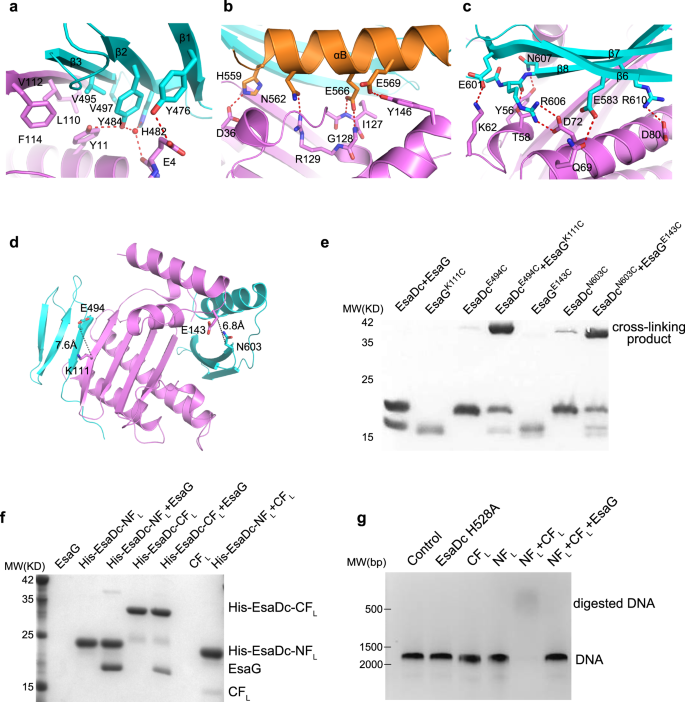
All of the secondary construction parts of EsaDc are concerned within the direct affiliation with EsaG, primarily by hydrogen bonds, salt bridges and hydrophobic packing interactions (Fig. 2a–c). Within the NF binding floor, residue Y476EsaDc on β1-strand kinds a hydrogen bond with the aspect chain of residue E4EsaG. The carbonyl oxygen of E4EsaG engages two water-mediated hydrogen bonds with Y484EsaDc and H482EsaDc alone the β2-strand, respectively. Moreover, Y484EsaDc donates a second hydrogen bond to Y11EsaG. Furthermore, residues on the β3-strand, together with V495EsaDc and V497EsaDc, type a hydrophobic core with the aspect chains of Y11, L110, V112 and F114 of EsaG (Fig. 2a).
a–c Shut-up view of the NF–EsaG (a), αB–EsaG (b) and the β-sheet (in CF)–EsaG (c) interactions. The proteins are coloured in the identical scheme as proven in Fig. 1a. The aspect chains of the interface residues are labeled and proven as sticks. The hydrogen bonds are depicted as pink dashed traces, and the water molecules are proven as pink spheres. d Pairs chosen for cross-linking assay. EsaGK111 and EsaDcE494 are chosen on the NF-EsaG interface. The closest distance between Cβ of EsaGK111 and EsaDcE494 is 7.6 Ǻ; EsaGE143 and EsaDcN603 are chosen near the CF-EsaG interface. The closest distance between the atoms on the aspect chains of EsaGE143 and EsaDcN603 is 6.8 Ǻ. e SDS-PAGE evaluation of EsaG and EsaDc variants incubated with the chemical probe. The cross-linked product has been labeled. f His-tag pull down assay. All of the proteins had been purified from E. Coli. A His-SUMO tag was co-expressed on the N-terminus of EsaDc-NFL (His-EsaDc-NFL) and EsaDc-NFL (His- EsaDc-CFL) to chelate with the nickel beads. Proteins are indicated by Coomassie blue staining. g in vitro nuclease exercise of NT-CT advanced. The gel was stained with ethidium bromide and visualized underneath UV gentle. The experiments had been carried out 3 times with comparable outcomes.
The CF binding is contributed collectively by αB and β6-8 of EsaDc, burying a solvent-accessible floor space of 3825 Å2, which is far bigger than that of the 2461 Å2 within the NF binding web site. Briefly, αB makes a hydrogen bond community to αD’ and αE’ of EsaG (Fig. 2b). The acidic aspect chain of E566EsaDc kinds a bidentate interplay with the spine amino teams of I127EsaG and G128EsaG on αD’ on the similar time. αD’ kinds a 3rd hydrogen bond with αB by the aspect chains of R129EsaG and N562EsaDc. Along with these interactions, αB is additional acknowledged by Y146EsaG on αE’ and D36EsaG on β1’. On the opposite aspect of the central β-sheet of EsaG, the β6-8 sheet is accommodated primarily by αB’ and αC’ of EsaG (Fig. 2c). β6 donates one hydrogen bond by E583EsaDc to Q69EsaG on αC’. E601 on β7 kinds a salt bridge with K62EsaG. On β8, two Arg residues (R606 and R610) type salt bridges with two Asp residues on αC’ (D72 and D80). Moreover, the spine carbonyl group of R606 and the aspect chain of N607 type hydrogen bond with T58EsaG and Y56EsaG from αB’, respectively.
Validation of EsaDc-EsaG interactions
To check whether or not EsaDc interacts with EsaG by two separated fragments in answer as noticed within the construction, we first carried out bio-layer interferometry (BLI) experiments (Supplementary Fig. 5). Substitution of Y484EsaDc from the NF-EsaG interface with an alanine diminished the binding of EsaDc and EsaG to a Okayd of 1.67 µM, about 3 times decrease than that of the wild-type (Okayd = 0.46 µM). On the CF-EsaG interface, mutant N562F barely decreased the binding affinity to a Okayd of 0.86 µM.
Cross-linking assays had been subsequent carried out to additional affirm the 2 interfaces. We chosen pairs of residues surrounding the interfaces for which Cβ–Cβ distances had been lower than 15 Å. These residues needs to be accessible to the solvent and never positioned in versatile areas. Based mostly on these standards, we recognized the pairs EsaGK111– EsaDcE494 across the NF-EsaG interface and EsaGE143– EsaDcN603 across the CF-EsaG interface (Fig. 2nd). We then launched cysteine residues at these positions and incubated the ensuing complexes with a chemical cross-linking probe. The probe incorporates two electrophilic iodoacetamide (IA) teams which can be separated by a 4-carbon spacer arm (Supplementary Fig. 6). IA probe may selectively goal and irreversibly cross-link cysteine in native proteins31. As proven in Fig. 2e, EsaG or EsaDc variants alone generated undetectable to week cross-linking alerts, however the alerts had been drastically elevated in each the pairs EsaGK111C-EsaDcE494C and EsaGE143C-EsaDcN603C. The cross-linked proteins had been recovered from SDS-PAGE, digested by trypsin and analyzed by liquid chromatography-MS (LC-MS) experiments. We efficiently recognized the anticipated cross-linked peptides in each samples as dominant cross-linked peptides (Supplementary Fig. 7 and Supplementary Knowledge 1), supporting that the residues of chosen pairs are in proximity as noticed within the construction.
We additionally carried out a His-tag pull-down assay to research the interplay of EsaG with every of the 2 fragments (Fig. 2f). We generated the 2 fragments of EsaDc in longer variations: the NF plus the lacking loop previous it (NFL, residues 450–503), and the CF plus the disordered ββα-metal finger motif (CFL, residues 504–614). The 2 fragments had been overexpressed with an N-terminal connected His-SUMO tag and known as His-EsaDc-NFL and His-EsaDc-CFL, respectively. As proven in Fig. 2f, each His-EsaDc-NFL and His-EsaDc-CFL can pull down EsaG effectively. Constantly, gel-filtration evaluation confirmed that NFL and CFL may type secure complexes and eluted with EsaG, respectively (Supplementary Fig. 8). Collectively, these outcomes validate the insertion of EsaG into the central β-sheet of EsaDc and that the 2 fragments of EsaDc generated throughout the insertion may work together with EsaG independently.
NFL and CFL reassemble into an lively protein within the absence of EsaG
Through the secretion course of, EsaG is left behind within the cell, whereas EsaD is transported to surrounding surroundings. We thus speculated that the NFL and CFL may reassemble into an intact construction of EsaDc when EsaG is faraway from the advanced. Within the His-tag pull-down assay, we noticed that CFL associates with the His-tagged NFL (Fig. 2f), indicative of formation of intact EsaDc protein. Additional proof for this concept comes from the NMR experiments. We collected two-dimensional 1H,15N-HSQC spectra for 15N-labeled NFL within the presence of unlabeled CFL, 15N-labeled CFL within the presence of unlabeled NFL and 15N-labeled EsaDc, respectively (Supplementary Fig. 9). Most alerts of the 15N-labeled NFL and CFL may overlap with the corresponding alerts of EsaDc, supporting that the NFL-CFL advanced and EsaDc share the identical construction. To check whether or not the reassembled construction is catalytically lively, we carried out an enzymatic assay utilizing a double-stranded DNA (dsDNA) as substrate (Fig. 2g). The outcomes confirmed that the NFL-CFL may digest DNA effectively within the presence of Mg2+, whereas neither NFL nor CFL alone exhibited any exercise. When EsaG was supplemented into the response, the nuclease exercise of NFL-CFL was completely abolished. These outcomes help that, throughout the secretion course of by T7SS, the covalently linked NFL and CFL can reassemble into an lively protein when EsaG is stripped off from EsaD-EsaG advanced. In truth, many proteins could be cut up into fragments that reassemble right into a practical protein spontaneously, similar to ribonuclease S and inexperienced fluorescent proteins32. Nonetheless, most of those fragments are generated by synthetic design, and their reassembly and disassembly will not be concerned within the practical regulation of those proteins within the nature.
Finger loop is the set off of conformational modifications on EsaDc
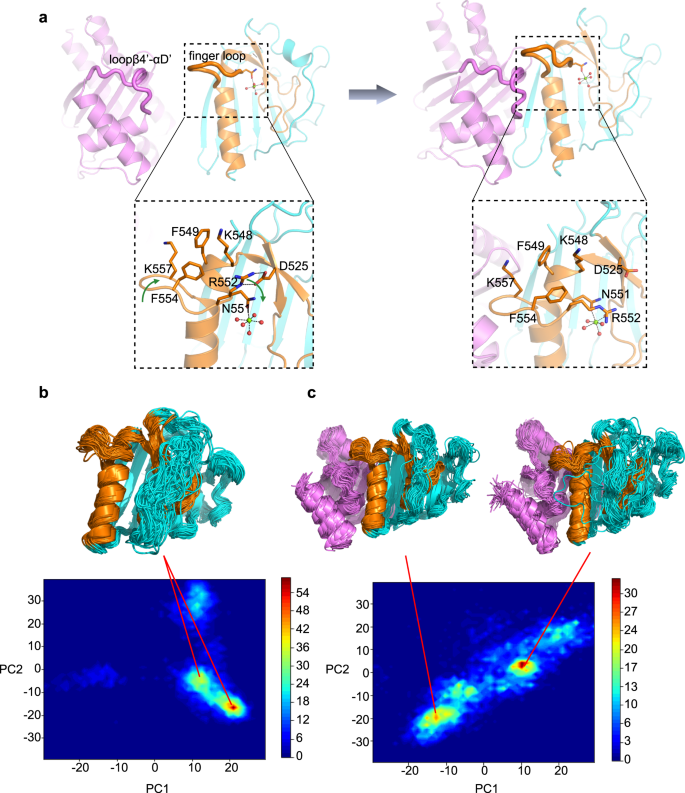
We subsequent investigated how EsaG triggers the conformational modifications of EsaDc. Discover that the EsaG binding floor of the CFL a part of the free EsaDc is uncovered to the solvent, whereas that of the NFL half is buried and stabilized by the ββα-metal finger motif (Fig. 1). It’s attainable that EsaG binds with the CFL first and triggers conformational change on the ββα-metal finger motif, resulting in dislocation of NFL from CFL. Structural superposition of the free and the EsaG-bound EsaDc by their CFL signifies steric conflict between the finger loop on the free EsaDc and Loopβ4’-αD’ of EsaG within the advanced (Supplementary Fig. 10).
For the reason that organic perform of the finger loop stays unknown, we first explored its roles in substrate binding and DNA cleavage. 4 residues (FKEK, 554–557) on the flip of the finger loop had been truncated. The ensuing constructs (EsaDc-FKEK and CFL-FKEK) exhibited comparable expression stage and conduct on chromatographic column to that of the full-length ones, suggesting that the truncation doesn’t destroy the entire construction of the proteins. Electrophoretic mobility shift assay (EMSA) confirmed that EsaDc-FKEK retains sturdy DNA binding capability (Supplementary Fig. 11a). In contrast to the EsaDc-DNA interplay that could possibly be whole abolished by EsaG, EsaDc-FKEK-DNA interplay nonetheless could possibly be noticed within the presence of EsaG, suggesting that the impact of EsaG to the ββα-metal finger motif was weakened by the truncation of finger loop. Nonetheless, the DNase exercise of NFL-CFL-FKEK advanced was drastically diminished and could possibly be additional diminished by EsaG (Supplementary Fig. 11b). These outcomes indicated that the finger loop just isn’t an important structural ingredient for DNA binding, however would possibly contain within the catalytic course of, in all probability by stabilizing the catalytic heart. Within the free EsaDc construction, we certainly noticed associations of the finger loop with residues surrounding the catalytic heart: R552, F554 and K557 from the finger loop type a hydrophobic area with K548 and F549 positioned on the αB which embraces the Mg2+ ion-coordinating residue N551; R552 additional contributes two hydrogen bonds with the aspect chain of D525 from β4’ (Fig. 3a).
a Conformational modifications of finger loop throughout the binding technique of EsaG to EsaDc. EsaG is coloured in purple; the ββα-metal finger is coloured in brown and the remainder a part of EsaDc in cyan. The residues on and across the finger loop are highlighted in expanded view with their aspect chains proven as stick presentation. The magnesium ion is proven as inexperienced sphere, and the coordinated waters are proven as pink spheres. b, c PCA evaluation for chance distribution of REST2 sampled EsaDc conformations in absence (b) or presence (c) of EsaG. The consultant buildings from excessive distributed reputation are proven in cartoon.
We then carried out molecular dynamics (MD) simulations together of ratchet&pawl potential33 to simulate the method of EsaG-EsaDc interplay. Ranging from EsaDc and EsaG separated, the ratchet coordinate was outlined because the RMSD to the construction of EsaDc-EsaG advanced, and bias potentials had been solely damped when the system tried to maneuver in the other way. No power was utilized when EsaG was spontaneously transferring in direction of EsaDc. Through the course of, it was seen that the finger loop was twisted as a result of approaching of Loopβ4’-αD’ on EsaG. F554 within the hydrophobic area moved inward to push R552, ensuing the damaged of the hydrogen bonds between R552 and D525. It’s attainable that the binding of Loopβ4’-αD’ on EsaG triggered the conformational modifications of the finger loop, which can deconstruct the hydrophobic and salt bridge interactions that keep the construction of ββα-metal finger motif on EsaDc (Fig. 3a).
To additional examine the method of EsaG-EsaDc interplay, we carried out reproduction trade with solute tempering (REST2) simulations34. The conformational area of EsaDc was sampled utilizing 16 replicas with efficient solute temperatures from 300 to 450 Okay. As a comparability, EsaDc in absence of EsaG was simulated underneath the identical situation. The simulation time was set to 500 ns for every reproduction, ensuing a whole sampling time of 8 μs for EsaDc and EsaDc-EsaG advanced, respectively. The conformation of EsaDc and its modifications induced by EsaG had been visualized utilizing principal part evaluation (PCA) based mostly on spine of EsaDc. As proven in Fig. 3b, c, the binding of EsaG considerably modified the conformational distributions of EsaDc. A number of new populations emerged, indicating the incidence of conformational heterogeneity upon binding to EsaG. The buildings of the populations had been examined and confirmed that though the NFL and CFL fragments didn’t separate within the time-limited sampling, the newly emerged buildings of the 2 fragments had been considerably extra disordered. Particularly, the ββα-metal finger motif within the presence of EsaG introduced as extremely versatile conformations. Collectively, the computational research revealed that the binding of EsaG induce conformational modifications of the finger loop first, which additional results in the dysfunction of the entire ββα-metal finger for the insertion mechanism.
EsaDc-EsaG type heterotetramer in answer
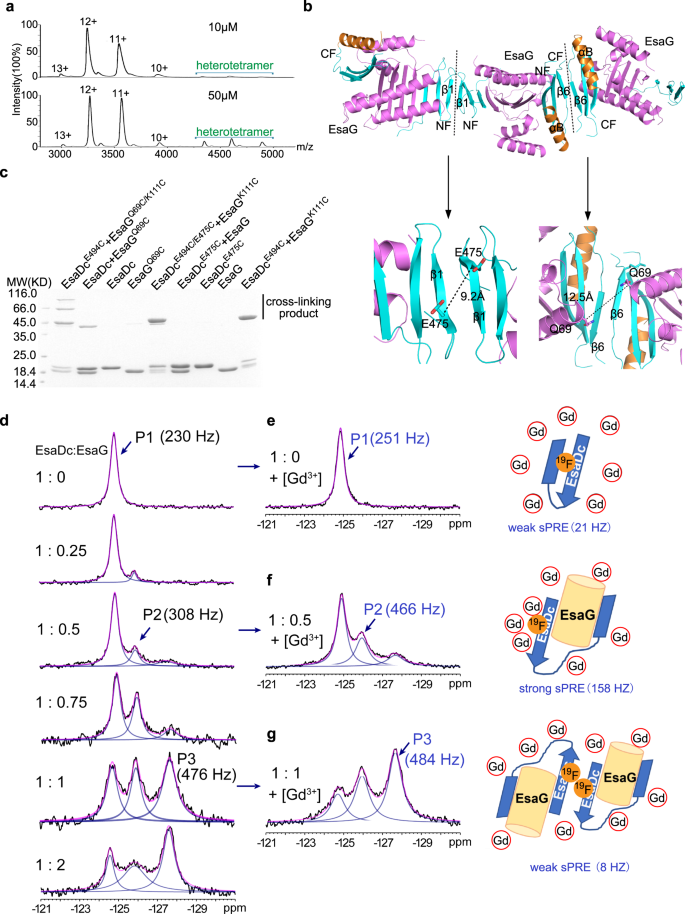
Despite the fact that there is just one EsaDc-EsaG advanced within the uneven unit, gel-filtration experiments confirmed that the advanced was eluted at a quantity akin to a molecular weight of 80 KD, near that of two EsaDc-EsaG dimers (Supplementary Fig. 1). We additional carried out native mass spectrometry to research the presence of the tetramer state of EsaDc-EsaG advanced. It’s obvious that the oligomeric state of the EsaDc-EsaG advanced exists and is focus dependent (Fig. 4a). At low focus (10 µM), the dominant oligomer in answer is EsaDc-EsaG dimer. When the focus is elevated to 50 µM, the alerts for tetramer are drastically strengthened.
a Native mass spectrum of EsaDc-EsaG advanced at low (10 µM) and excessive (50 µM) concentrations. Experimental molecular weights for dimer and tetramer calculated based mostly on the assigned cost states are 39.092 KD (theoretical molecular weight 38.764 KD) and 77.789 KD (theoretical molecular weight 77.528 KD), respectively. b Ribbon illustration of EsaDc-EsaG heterodimer interacting with close by uneven items. The proteins are coloured in the identical scheme as proven in Fig. 1a. Potential dimer-to-dimer interfaces throughout the crystal unit cells are introduced as black sprint. Shut-up view of the residues chosen for cross-linking are highlighted under. c SDS-PAGE evaluation of EsaG and EsaDc variants incubated with the IA probe. The experiments had been carried out twice with comparable outcomes. d 19F-NMR spectra of EsaDc-19F-W567 within the presence of various equivalents of EsaG. Black traces are the experimental spectra, blue traces are deconvoluted peaks and magenta traces are the sum of deconvoluted peaks. e–g are the spectra (left) of EsaDc19F-W567 blended with the indicated equal of EsaG and 50 mM Ga-DTPA-BMA. The cartoon representations (proper) are the solvent accessibility of the 19F-W567 in free EsaDc, akin to P1 (e), in EsaDc-EsaG dimer, akin to P2 (f), and in EsaDc-EsaG tetramer, akin to P3 (g), respectively. The sPRE strengths in Hz are indicated underneath the cartoons.
To disclose how the EsaDc-EsaG tetramer is shaped, we subsequent searched the heterotetramer interface throughout the crystal items. There are two crystallographic packing contacts probably concerned in tetramer formation, each of that are generated by two of the identical fragment of EsaDc in a 2-fold symmetry, whereas EsaG doesn’t contribute any direct interplay (Fig. 4b). Within the first contact, three-stranded β-sheets of two NFs work together by their anti-paralleled β1, whereas the second is constructed by αB’ and β6’ from two CFs (Fig. 4b). To find out which interface mediates the tetramer formation, we evaluated the cross-linking of residue EsaDcE475C, positioned on the NF-mediated interface, and EsaGQ69C, positioned on the CF-mediated interface, utilizing IA probe (Fig. 4b). The outcomes confirmed that sturdy sign of cross-linked product was generated solely by EsaGQ69C within the presence of EsaDc, suggesting that two EsaDc-EsaG dimers type a tetramer by the CFs (Fig. 4c). We additionally mixed these two mutations with the pair EsaGK111C– EsaDcE494C on the NF-EsaG interface, respectively. As anticipated, the EsaGQ69C/K111C– EsaDcE494C advanced generated cross-linked merchandise of trimer (~60 KD) and tetramer (~80 KD), whereas the EsaGK111C-EsaDcE494C/E475C advanced solely exhibited a cross-linked dimer, as that of the pair EsaGK111C– EsaDcE494C (Fig. 4c).
Interplay of EsaDc and EsaG and the interface for the tetramer formation had been additional confirmed by utilizing answer fluorine NMR. There are two tryptophan residues in EsaDc (W560 and W567), each of that are buried within the free EsaDc construction, however uncovered to solvent on the construction of the advanced (Supplementary Fig. 12). The C5-position of W567 is pointing in direction of the solvent, making it an appropriate goal to be substituted with 5-fluoro-L-tryptophan (5F-L-Trp) and to review the conformational modifications of EsaDc by 19F NMR. To simplify the NMR spectrum, W560 was mutated to phenylalanine. The resulted single web site 19F labeled EsaDc (EsaDc-19F-W567) confirmed a single peak (P1) at −124.8 ppm within the 19F NMR spectrum. Upon addition of EsaG, a brand new peak (P2) appeared at −125.9 ppm. With the rise of EsaG, a 3rd peak (P3) appeared at −127.6 ppm and have become the dominant peak (Fig. 4d). The sequential look of P2 and P3 indicated that the binding of EsaDc and EsaG just isn’t a single course of. Contemplating a big change of the solvent accessibility of W567 based mostly on crystal buildings (Supplementary Fig. 12b), solvent paramagnetic resonance enhancement (sPRE) measurement35 was carried out to evaluate the solvent publicity of 19F-W567. Upon addition of a paramagnetic reagent gadodiamine (Gd-DTPA-BMA), which may broaden the resonance peaks for nucleic spins with shut contact, the road width of P1 was solely barely modified, per a buried 19F-W567 in EsaDc (Fig. 4e). In distinction, within the presence of EsaG, the newly appeared P2 confirmed a powerful sPRE impact (158 Hz), indicating a solvent uncovered surroundings of 19F-W567 which permits the paramagnetic Gd-DTPA-BMA method with brief distance. That is per the crystal construction of the EsaDc-EsaG advanced the place W567 is uncovered to solvent (Fig. 4f and Supplementary Fig. 12b). Surprisingly, P3, which is very populated within the presence of excessive focus of EsaG, didn’t present important sPRE impact (Fig. 4g). We rationalize that P3 corresponds to the formation of EsaDc-EsaG tetramer, the place W567 is positioned in a pocket comprised αB’ and β6’ from two CFs (Fig. 4f and Supplementary Fig. 12c). The pocket has Van der Waals diameter of <6 Å and is smaller than the paramagnetic reagent Gd-DTPA-BMA (vdW diameter: >8 Å)36, thus, stopping an in depth contact between Gd-DTPA-BMA and 19F-W567 and leading to a weak PRE impact. Total, the sequential look of P2 and P3 in addition to the sturdy sPRE for P2 and weak sPRE for P3 are per the structural mannequin of EsaDc-EsaG dimer and EsaDc-EsaG tetramer.
A conserved inhibition mechanism in Gram-positive micro organism
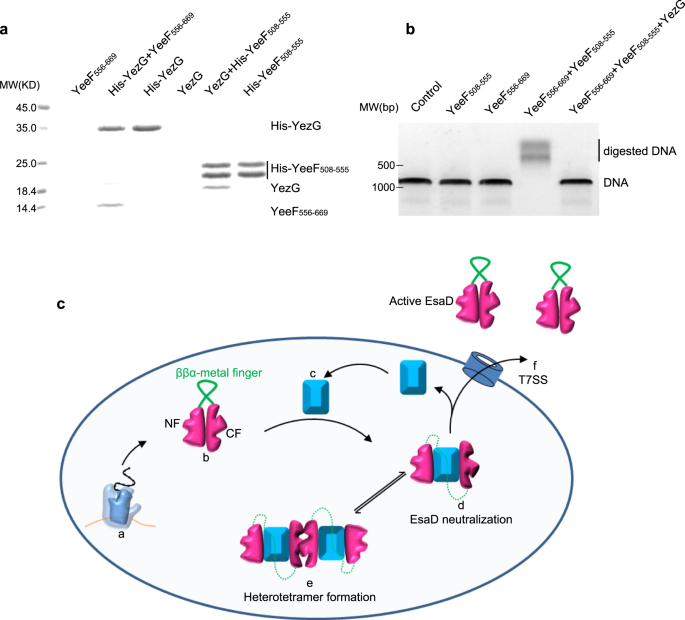
Sequence blast evaluation recognized extra EsaG-like proteins in lots of different Gram-positive micro organism (Supplementary Fig. 13), sharing about 40% sequence similarity with EsaG. All these proteins are categorized right into a household named DUF600 (Area of Unknown Perform), resulting from restricted information about their organic perform. One exception is YezG from Bacillus subtilis, top-of-the-line characterised micro organism and normally used as a mannequin organism for Gram-positive micro organism. Much like EsaG, YezG interacts with and inhibits the exercise of a nuclease area positioned on the C-terminus of a toxin, YeeF37. To analyze whether or not YezG inhibits YeeF utilizing an identical technique, we cleaved the nuclease area of YeeF into two fragments (YeeF508-555 and YeeF556-669) in keeping with the sequences of NTL and CTL of EsaDc. As is the case for EsaD, the 2 fragments of YeeF may work together with YezG independently within the His-tag pull-down assay (Fig. 5a). They might additionally assemble into lively type protein to digest dsDNA, however this exercise is inhibited by YezG (Fig. 5b). These outcomes indicate that the inhibition of nuclease toxin by insertion of antitoxin into the core β-sheet of the toxin and deformation of the ββα-metal finger motif could also be extensively adopted amongst Gram-positive micro organism.
a His-tag pull down assay. A His-SUMO tag was co-expressed on the N-terminus of YezG (His-YezG) and YeeF508-555 (His-YeeF508-555) to chelate with the nickel beads. All of the proteins had been purified from E. Coli. Proteins are indicated by Coomassie blue staining. The experiments had been carried out twice with comparable outcomes. b In vitro nuclease exercise of YeeF fragments. The gel was stained with ethidium bromide and visualized underneath UV gentle. The experiments had been carried out 3 times with comparable outcomes. c Cartoon illustration of the molecular mannequin for the mechanism of EsaD inhibition by EsaG. The nuclease exercise of EsaD is inhibited upon its synthesis by insertion of EsaG monomer into the core construction of EsaD, resulting in the dysfunction of the ββα-metal finger. The EsaD-EsaG advanced could type tetramer when amassed within the micro organism. EsaG proteins are faraway from the advanced and left behind when EsaD proteins are translocated by the T7SS. This mannequin exhibits the ribosome (a), the newly synthetized EsaD (b), EsaG (c), EsaD-EsaG heterodimer (d), EsaD-EsaG heterotetramer (e) and the T7SS (f).